bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0714_orf1
Length=96
Score E
Sequences producing significant alignments: (Bits) Value
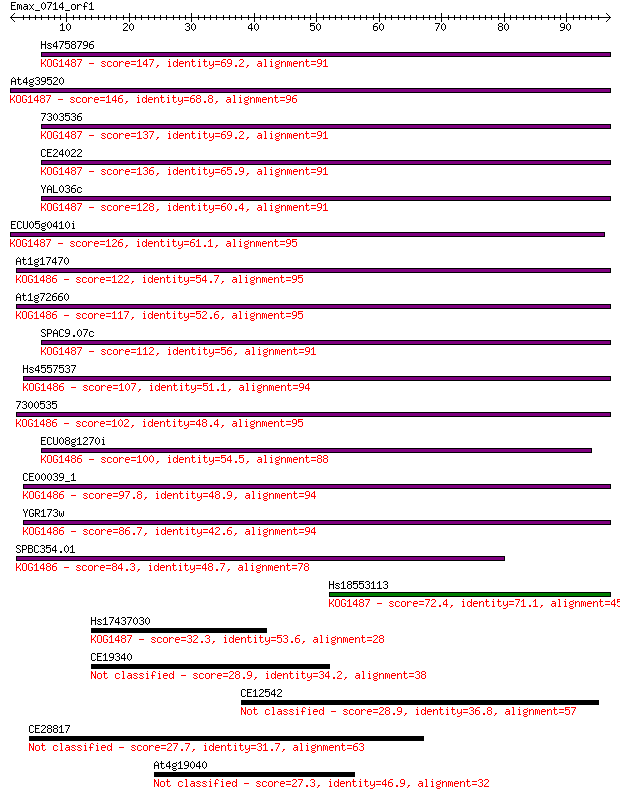
Hs4758796 147 5e-36
At4g39520 146 1e-35
7303536 137 7e-33
CE24022 136 9e-33
YAL036c 128 3e-30
ECU05g0410i 126 1e-29
At1g17470 122 2e-28
At1g72660 117 4e-27
SPAC9.07c 112 2e-25
Hs4557537 107 7e-24
7300535 102 2e-22
ECU08g1270i 100 9e-22
CE00039_1 97.8 5e-21
YGR173w 86.7 9e-18
SPBC354.01 84.3 5e-17
Hs18553113 72.4 2e-13
Hs17437030 32.3 0.20
CE19340 28.9 2.3
CE12542 28.9 2.3
CE28817 27.7 5.2
At4g19040 27.3 6.5
> Hs4758796
Length=367
Score = 147 bits (371), Expect = 5e-36, Method: Compositional matrix adjust.
Identities = 63/91 (69%), Positives = 78/91 (85%), Gaps = 0/91 (0%)
Query 6 NLDGLLEKIWEYLSLIRVYTKPKGQIPDYNAPVILSSGRCKIEDFCLKIHKSILNEFKQA 65
N D LLEKIW+YL L+R+YTKPKGQ+PDY +PV+L R +EDFC+KIHK+++ EFK A
Sbjct 277 NFDDLLEKIWDYLKLVRIYTKPKGQLPDYTSPVVLPYSRTTVEDFCMKIHKNLIKEFKYA 336
Query 66 IVWGKSVKHNPQKVGKEHQLEDEDVVQIIKK 96
+VWG SVKHNPQKVGK+H LEDEDV+QI+KK
Sbjct 337 LVWGLSVKHNPQKVGKDHTLEDEDVIQIVKK 367
> At4g39520
Length=369
Score = 146 bits (368), Expect = 1e-35, Method: Compositional matrix adjust.
Identities = 66/96 (68%), Positives = 80/96 (83%), Gaps = 0/96 (0%)
Query 1 ANLELNLDGLLEKIWEYLSLIRVYTKPKGQIPDYNAPVILSSGRCKIEDFCLKIHKSILN 60
A+LE NLDGLL+KIWEYL L R+YTKPK PDY+ PVILSS + +EDFC++IHK +L
Sbjct 273 AHLEWNLDGLLDKIWEYLDLTRIYTKPKAMNPDYDDPVILSSKKRTVEDFCIRIHKDMLK 332
Query 61 EFKQAIVWGKSVKHNPQKVGKEHQLEDEDVVQIIKK 96
+FK A+VWG S KH PQ+VGKEH+LEDEDVVQI+KK
Sbjct 333 QFKYALVWGSSAKHKPQRVGKEHELEDEDVVQIVKK 368
> 7303536
Length=368
Score = 137 bits (344), Expect = 7e-33, Method: Compositional matrix adjust.
Identities = 63/91 (69%), Positives = 73/91 (80%), Gaps = 0/91 (0%)
Query 6 NLDGLLEKIWEYLSLIRVYTKPKGQIPDYNAPVILSSGRCKIEDFCLKIHKSILNEFKQA 65
N D LLE +WEYL L R+YTKPKGQ+PDYN+PV+L + R IEDFC K+H+SI EFK A
Sbjct 277 NFDDLLELMWEYLRLQRIYTKPKGQLPDYNSPVVLHNERTSIEDFCNKLHRSIAKEFKYA 336
Query 66 IVWGKSVKHNPQKVGKEHQLEDEDVVQIIKK 96
+VWG SVKH PQKVG EH L DEDVVQI+KK
Sbjct 337 LVWGSSVKHQPQKVGIEHVLNDEDVVQIVKK 367
> CE24022
Length=366
Score = 136 bits (343), Expect = 9e-33, Method: Compositional matrix adjust.
Identities = 60/91 (65%), Positives = 76/91 (83%), Gaps = 0/91 (0%)
Query 6 NLDGLLEKIWEYLSLIRVYTKPKGQIPDYNAPVILSSGRCKIEDFCLKIHKSILNEFKQA 65
N D LLEK+WEYL+LIR+YTKPKGQ+PDY+ P++L++ R IED C KIHKS+ +FK A
Sbjct 275 NFDDLLEKVWEYLNLIRIYTKPKGQLPDYSQPIVLNAERKSIEDLCTKIHKSLQKDFKCA 334
Query 66 IVWGKSVKHNPQKVGKEHQLEDEDVVQIIKK 96
+VWG S KHNPQ+VG++H L DEDVVQ+IKK
Sbjct 335 LVWGASAKHNPQRVGRDHVLIDEDVVQVIKK 365
> YAL036c
Length=369
Score = 128 bits (321), Expect = 3e-30, Method: Compositional matrix adjust.
Identities = 55/91 (60%), Positives = 74/91 (81%), Gaps = 0/91 (0%)
Query 6 NLDGLLEKIWEYLSLIRVYTKPKGQIPDYNAPVILSSGRCKIEDFCLKIHKSILNEFKQA 65
NLD LL+ +W+ L+L+R+YTKPKGQIPD+ PV+L S RC ++DFC +IHKS++++F+ A
Sbjct 279 NLDELLQVMWDRLNLVRIYTKPKGQIPDFTDPVVLRSDRCSVKDFCNQIHKSLVDDFRNA 338
Query 66 IVWGKSVKHNPQKVGKEHQLEDEDVVQIIKK 96
+V+G SVKH PQ VG H LEDEDVV I+KK
Sbjct 339 LVYGSSVKHQPQYVGLSHILEDEDVVTILKK 369
> ECU05g0410i
Length=362
Score = 126 bits (316), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 58/95 (61%), Positives = 68/95 (71%), Gaps = 0/95 (0%)
Query 1 ANLELNLDGLLEKIWEYLSLIRVYTKPKGQIPDYNAPVILSSGRCKIEDFCLKIHKSILN 60
AN N D LL K+W YL L+R+Y KPKG+ DY PVIL S + + DFC IH+ IL
Sbjct 266 ANFGWNFDSLLSKMWSYLDLVRIYPKPKGEPIDYEEPVILRSNKRSVADFCNAIHRGILA 325
Query 61 EFKQAIVWGKSVKHNPQKVGKEHQLEDEDVVQIIK 95
+FK A+VWG SVKHNPQKVGKEH L D DVVQI+K
Sbjct 326 KFKHALVWGSSVKHNPQKVGKEHILNDSDVVQIVK 360
> At1g17470
Length=399
Score = 122 bits (306), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 52/97 (53%), Positives = 73/97 (75%), Gaps = 2/97 (2%)
Query 2 NLELNLDGLLEKIWEYLSLIRVYTKPKGQIPDYNAPVILSS--GRCKIEDFCLKIHKSIL 59
NL+LNLD LL ++W+ + L+RVY+KP+GQ PD++ P +LSS G C +EDFC +H++++
Sbjct 271 NLKLNLDRLLARMWDEMGLVRVYSKPQGQQPDFDEPFVLSSDRGGCTVEDFCNHVHRTLV 330
Query 60 NEFKQAIVWGKSVKHNPQKVGKEHQLEDEDVVQIIKK 96
+ K A+VWG S +HNPQ G LEDEDVVQI+KK
Sbjct 331 KDMKYALVWGTSTRHNPQNCGLSQHLEDEDVVQIVKK 367
> At1g72660
Length=399
Score = 117 bits (294), Expect = 4e-27, Method: Compositional matrix adjust.
Identities = 50/97 (51%), Positives = 73/97 (75%), Gaps = 2/97 (2%)
Query 2 NLELNLDGLLEKIWEYLSLIRVYTKPKGQIPDYNAPVILSS--GRCKIEDFCLKIHKSIL 59
NL+LNLD LL ++W+ + L+RVY+KP+ Q PD++ P +LS+ G C +EDFC ++H++++
Sbjct 271 NLKLNLDRLLARMWDEMGLVRVYSKPQSQQPDFDEPFVLSADRGGCTVEDFCNQVHRTLV 330
Query 60 NEFKQAIVWGKSVKHNPQKVGKEHQLEDEDVVQIIKK 96
+ K A+VWG S +H PQ G H LEDEDVVQI+KK
Sbjct 331 KDMKYALVWGTSARHYPQHCGLFHHLEDEDVVQIVKK 367
> SPAC9.07c
Length=366
Score = 112 bits (279), Expect = 2e-25, Method: Composition-based stats.
Identities = 51/91 (56%), Positives = 66/91 (72%), Gaps = 0/91 (0%)
Query 6 NLDGLLEKIWEYLSLIRVYTKPKGQIPDYNAPVILSSGRCKIEDFCLKIHKSILNEFKQA 65
N+D L E +W+YL+L+RVYT+P+G PDY+ PVIL +G +EDFC IH SI ++FK A
Sbjct 276 NIDELKETMWDYLNLVRVYTRPRGLEPDYSEPVILRTGHSTVEDFCNNIHSSIKSQFKHA 335
Query 66 IVWGKSVKHNPQKVGKEHQLEDEDVVQIIKK 96
VWGKSV + +VG H L DEDVV I+KK
Sbjct 336 YVWGKSVPYPGMRVGLSHVLLDEDVVTIVKK 366
> Hs4557537
Length=364
Score = 107 bits (266), Expect = 7e-24, Method: Composition-based stats.
Identities = 48/94 (51%), Positives = 69/94 (73%), Gaps = 1/94 (1%)
Query 3 LELNLDGLLEKIWEYLSLIRVYTKPKGQIPDYNAPVILSSGRCKIEDFCLKIHKSILNEF 62
++LNLD LLE +WEYL+L +YTK +GQ PD+ +IL G +E C +IH+S+ ++F
Sbjct 272 MKLNLDYLLEMLWEYLALTCIYTKKRGQRPDFTDAIILRKG-ASVEHVCHRIHRSLASQF 330
Query 63 KQAIVWGKSVKHNPQKVGKEHQLEDEDVVQIIKK 96
K A+VWG S K++PQ+VG H +E EDV+QI+KK
Sbjct 331 KYALVWGTSTKYSPQRVGLTHTMEHEDVIQIVKK 364
> 7300535
Length=363
Score = 102 bits (253), Expect = 2e-22, Method: Composition-based stats.
Identities = 46/95 (48%), Positives = 67/95 (70%), Gaps = 1/95 (1%)
Query 2 NLELNLDGLLEKIWEYLSLIRVYTKPKGQIPDYNAPVILSSGRCKIEDFCLKIHKSILNE 61
N++LNLD ++E +WE L LIRVYTK G PD++ +IL G +E C IH+++ +
Sbjct 270 NMKLNLDYMMEALWEALQLIRVYTKKPGAPPDFDDGLILRKG-VSVEHVCHAIHRTLAAQ 328
Query 62 FKQAIVWGKSVKHNPQKVGKEHQLEDEDVVQIIKK 96
FK A+VWG S K++PQ+VG H + DEDV+Q++KK
Sbjct 329 FKYALVWGTSTKYSPQRVGIAHVMADEDVIQVVKK 363
> ECU08g1270i
Length=362
Score = 100 bits (248), Expect = 9e-22, Method: Composition-based stats.
Identities = 48/88 (54%), Positives = 59/88 (67%), Gaps = 1/88 (1%)
Query 6 NLDGLLEKIWEYLSLIRVYTKPKGQIPDYNAPVILSSGRCKIEDFCLKIHKSILNEFKQA 65
N+ LLE IW+ L L RVYTK KG P + PV++ G ++D C +IHK L FK A
Sbjct 273 NISRLLEDIWDKLKLTRVYTKKKGAFPSLDDPVVIRKGG-TVKDLCSRIHKDFLLSFKHA 331
Query 66 IVWGKSVKHNPQKVGKEHQLEDEDVVQI 93
+VWG S KH+PQ+VG H LEDEDVVQI
Sbjct 332 LVWGTSAKHSPQRVGLGHTLEDEDVVQI 359
> CE00039_1
Length=408
Score = 97.8 bits (242), Expect = 5e-21, Method: Compositional matrix adjust.
Identities = 46/95 (48%), Positives = 64/95 (67%), Gaps = 1/95 (1%)
Query 3 LELNLDGLLEKIWEYLSLIRVYTKPKGQIPDYNAP-VILSSGRCKIEDFCLKIHKSILNE 61
+ LN+D LLEK+WEYL+L+RVYTK G PD I+ G IE C +H+SI +
Sbjct 272 MNLNMDYLLEKMWEYLALVRVYTKKPGNAPDLGPEDGIILRGGATIEHCCHALHRSIAAQ 331
Query 62 FKQAIVWGKSVKHNPQKVGKEHQLEDEDVVQIIKK 96
+ AIVWG S K +PQ+VG H+L+ EDV+QI+++
Sbjct 332 LRYAIVWGTSTKFSPQRVGLHHKLDHEDVIQIMRR 366
> YGR173w
Length=368
Score = 86.7 bits (213), Expect = 9e-18, Method: Compositional matrix adjust.
Identities = 40/94 (42%), Positives = 62/94 (65%), Gaps = 1/94 (1%)
Query 3 LELNLDGLLEKIWEYLSLIRVYTKPKGQIPDYNAPVILSSGRCKIEDFCLKIHKSILNEF 62
++L L ++E+IW L+L RVYTK +G P ++ P+++ + I D C IH+ ++F
Sbjct 276 MDLGLQDVVEEIWYQLNLSRVYTKKRGVRPVFDDPLVVRNN-STIGDLCHGIHRDFKDKF 334
Query 63 KQAIVWGKSVKHNPQKVGKEHQLEDEDVVQIIKK 96
K A+VWG S KH+PQK G H+++DEDVV + K
Sbjct 335 KYALVWGSSAKHSPQKCGLNHRIDDEDVVSLFAK 368
> SPBC354.01
Length=346
Score = 84.3 bits (207), Expect = 5e-17, Method: Composition-based stats.
Identities = 38/78 (48%), Positives = 56/78 (71%), Gaps = 1/78 (1%)
Query 2 NLELNLDGLLEKIWEYLSLIRVYTKPKGQIPDYNAPVILSSGRCKIEDFCLKIHKSILNE 61
N++LNLD L E+IWE L+L R+YTK KG++PD++ +I+ G IE C +IH+++ +
Sbjct 270 NMKLNLDFLKERIWEELNLYRIYTKRKGEMPDFSEALIVRKGS-TIEQVCNRIHRTLAEQ 328
Query 62 FKQAIVWGKSVKHNPQKV 79
K A+VWG S KH+PQ V
Sbjct 329 LKYALVWGTSAKHSPQVV 346
> Hs18553113
Length=94
Score = 72.4 bits (176), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 32/45 (71%), Positives = 41/45 (91%), Gaps = 0/45 (0%)
Query 52 LKIHKSILNEFKQAIVWGKSVKHNPQKVGKEHQLEDEDVVQIIKK 96
+KIHK+++ EFK A+VWG SVKHNPQKVGK+H LED+DV+QI+KK
Sbjct 1 MKIHKNLIKEFKYALVWGLSVKHNPQKVGKDHTLEDKDVIQIVKK 45
> Hs17437030
Length=104
Score = 32.3 bits (72), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 15/28 (53%), Positives = 20/28 (71%), Gaps = 0/28 (0%)
Query 14 IWEYLSLIRVYTKPKGQIPDYNAPVILS 41
I E LSL++VYT PKGQ + + V+LS
Sbjct 76 IGEDLSLVKVYTNPKGQPQNCTSLVVLS 103
> CE19340
Length=354
Score = 28.9 bits (63), Expect = 2.3, Method: Composition-based stats.
Identities = 13/42 (30%), Positives = 24/42 (57%), Gaps = 4/42 (9%)
Query 14 IWEYLSLIRVYTKPKGQIPDYNAPVILSSGRCKI----EDFC 51
++E+ +YTK + Q+ D+N+ + ++ CK EDFC
Sbjct 24 VFEFTEFEFLYTKSQFQVEDFNSSLPSTAENCKCDTVNEDFC 65
> CE12542
Length=399
Score = 28.9 bits (63), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 21/63 (33%), Positives = 31/63 (49%), Gaps = 6/63 (9%)
Query 38 VILSSGRCKIE------DFCLKIHKSILNEFKQAIVWGKSVKHNPQKVGKEHQLEDEDVV 91
V+ G CKIE +F K+H +I + KQ KS N Q++G E+ + +
Sbjct 284 VLYFPGGCKIEKLLKLQNFHRKVHVNIFDALKQLPPSVKSWLENRQEIGTEYTFDISNED 343
Query 92 QII 94
QII
Sbjct 344 QII 346
> CE28817
Length=681
Score = 27.7 bits (60), Expect = 5.2, Method: Composition-based stats.
Identities = 20/63 (31%), Positives = 28/63 (44%), Gaps = 8/63 (12%)
Query 4 ELNLDGLLEKIWEYLSLIRVYTKPKGQIPDYNAPVILSSGRCKIEDFCLKIHKSILNEFK 63
+L D LE IW I V T P GQ + GR I+ C++I SI+ F
Sbjct 466 QLVFDFALESIWGKFHDITVDTAPFGQ-------CLTKHGR-SIDSLCVQIDDSIIEMFS 517
Query 64 QAI 66
+ +
Sbjct 518 EVL 520
> At4g19040
Length=679
Score = 27.3 bits (59), Expect = 6.5, Method: Composition-based stats.
Identities = 15/34 (44%), Positives = 19/34 (55%), Gaps = 4/34 (11%)
Query 24 YTKPKGQIPDYNAPV--ILSSGRCKIEDFCLKIH 55
Y K K Q DY P+ +L G C++ED LK H
Sbjct 37 YYKKKPQ--DYQVPIKTMLIDGNCRVEDRGLKTH 68
Lambda K H
0.318 0.138 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1201432980
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40