bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0680_orf2
Length=131
Score E
Sequences producing significant alignments: (Bits) Value
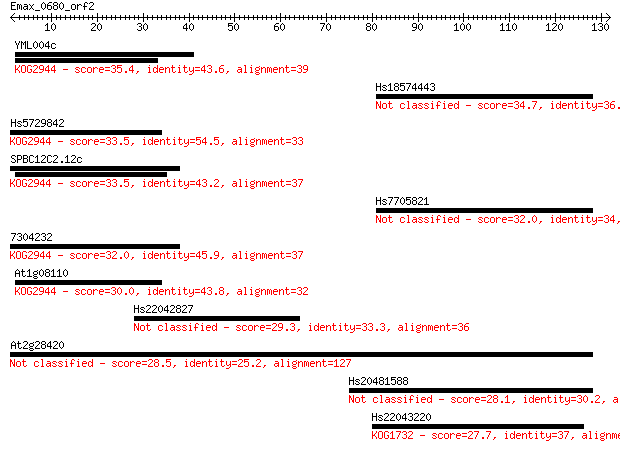
YML004c 35.4 0.030
Hs18574443 34.7 0.042
Hs5729842 33.5 0.10
SPBC12C2.12c 33.5 0.12
Hs7705821 32.0 0.34
7304232 32.0 0.35
At1g08110 30.0 1.1
Hs22042827 29.3 2.3
At2g28420 28.5 3.2
Hs20481588 28.1 5.0
Hs22043220 27.7 5.7
> YML004c
Length=326
Score = 35.4 bits (80), Expect = 0.030, Method: Compositional matrix adjust.
Identities = 17/39 (43%), Positives = 24/39 (61%), Gaps = 0/39 (0%)
Query 2 TLKFYEDVLGMTLIDKKTAGSFGFTLYFLALESPTSTSL 40
+L+FY++VLGM L+ S FTLYFL P + S+
Sbjct 196 SLEFYQNVLGMKLLRTSEHESAKFTLYFLGYGVPKTDSV 234
Score = 29.6 bits (65), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 13/31 (41%), Positives = 19/31 (61%), Gaps = 0/31 (0%)
Query 2 TLKFYEDVLGMTLIDKKTAGSFGFTLYFLAL 32
T+KFY + GM L+ +K F+LYFL+
Sbjct 36 TVKFYTEHFGMKLLSRKDFEEAKFSLYFLSF 66
> Hs18574443
Length=391
Score = 34.7 bits (78), Expect = 0.042, Method: Compositional matrix adjust.
Identities = 17/47 (36%), Positives = 32/47 (68%), Gaps = 3/47 (6%)
Query 81 FLCAESNEEKILKRIKRNYVKVELVDDEVYGQKVLLLRDPQNIPIRI 127
FLC E++ EK+LK+I R Y++ + +++++ + LLL DP +R+
Sbjct 340 FLCMETHYEKVLKKISR-YIQEQ--NEKIFAPRGLLLTDPVERGMRV 383
> Hs5729842
Length=184
Score = 33.5 bits (75), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 18/33 (54%), Positives = 21/33 (63%), Gaps = 0/33 (0%)
Query 1 ETLKFYEDVLGMTLIDKKTAGSFGFTLYFLALE 33
++L FY VLGMTLI K F+LYFLA E
Sbjct 44 KSLDFYTRVLGMTLIQKCDFPIMKFSLYFLAYE 76
> SPBC12C2.12c
Length=302
Score = 33.5 bits (75), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 16/37 (43%), Positives = 23/37 (62%), Gaps = 0/37 (0%)
Query 1 ETLKFYEDVLGMTLIDKKTAGSFGFTLYFLALESPTS 37
++LKFY +V GM LID+ F+L FLA + P +
Sbjct 24 KSLKFYTEVFGMKLIDQWVFEENEFSLSFLAFDGPGA 60
Score = 27.7 bits (60), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 16/33 (48%), Positives = 20/33 (60%), Gaps = 1/33 (3%)
Query 2 TLKFYEDVLGMTLIDKKTAGSFGFTLYFLALES 34
++ FYE LGM +IDK + FT YFLA S
Sbjct 180 SIAFYEK-LGMKVIDKADHPNGKFTNYFLAYPS 211
> Hs7705821
Length=104
Score = 32.0 bits (71), Expect = 0.34, Method: Compositional matrix adjust.
Identities = 16/47 (34%), Positives = 32/47 (68%), Gaps = 3/47 (6%)
Query 81 FLCAESNEEKILKRIKRNYVKVELVDDEVYGQKVLLLRDPQNIPIRI 127
FLC E++ EK+LK++ + Y++ + ++++Y + LLL DP +R+
Sbjct 46 FLCMETHYEKVLKKVSK-YIQEQ--NEKIYAPQGLLLTDPIERGLRV 89
> 7304232
Length=176
Score = 32.0 bits (71), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 17/37 (45%), Positives = 22/37 (59%), Gaps = 0/37 (0%)
Query 1 ETLKFYEDVLGMTLIDKKTAGSFGFTLYFLALESPTS 37
++L FY VLGMTL+ K F+LYFL E+ T
Sbjct 40 KSLPFYTGVLGMTLLVKLDFPEAKFSLYFLGYENATD 76
> At1g08110
Length=185
Score = 30.0 bits (66), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 14/32 (43%), Positives = 19/32 (59%), Gaps = 0/32 (0%)
Query 2 TLKFYEDVLGMTLIDKKTAGSFGFTLYFLALE 33
+L FY VLGM+L+ + F+LYFL E
Sbjct 41 SLDFYSRVLGMSLLKRLDFSEMKFSLYFLGYE 72
> Hs22042827
Length=522
Score = 29.3 bits (64), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 12/36 (33%), Positives = 19/36 (52%), Gaps = 0/36 (0%)
Query 28 YFLALESPTSTSLHYWLWLQRFSSICIKVHADGNHV 63
Y LA SP ++L YWLW ++ +++ N V
Sbjct 121 YLLAQTSPPESNLVYWLWEKQKVMAIVRIDTQNNPV 156
> At2g28420
Length=184
Score = 28.5 bits (62), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 32/129 (24%), Positives = 54/129 (41%), Gaps = 22/129 (17%)
Query 1 ETLKFYEDVLGMTLIDKKTAGSFGFTLYFLALESPTSTSLHYWLWLQRFSSI-CIKVHAD 59
++L+FY VLG I++ SF F +L +Y + + + K+ +D
Sbjct 33 KSLEFYTKVLGFVEIER--PASFDFDGAWL---------FNYGVGIHLVQAKDQDKLPSD 81
Query 60 GNHVAPYKDLGPSECGFLGISFLCAESNE-EKILKRIKRNYVKVELVDDEVYGQKVLLLR 118
+H+ P + ISF C + EK LK +K Y+K + D++ L
Sbjct 82 TDHLDPMDN---------HISFQCEDMEALEKRLKEVKVKYIKRTVGDEKDAAIDQLFFN 132
Query 119 DPQNIPIRI 127
DP + I
Sbjct 133 DPDGFMVEI 141
> Hs20481588
Length=321
Score = 28.1 bits (61), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 16/53 (30%), Positives = 26/53 (49%), Gaps = 0/53 (0%)
Query 75 GFLGISFLCAESNEEKILKRIKRNYVKVELVDDEVYGQKVLLLRDPQNIPIRI 127
G LG + S E K +RN + +L E +GQ+ +L R + +P R+
Sbjct 102 GILGREEILVNSVETKQCYSARRNDQEAQLGSLEKWGQRRILQRPDRRVPTRV 154
> Hs22043220
Length=667
Score = 27.7 bits (60), Expect = 5.7, Method: Composition-based stats.
Identities = 17/47 (36%), Positives = 26/47 (55%), Gaps = 2/47 (4%)
Query 80 SFL-CAESNEEKILKRIKRNYVKVELVDDEVYGQKVLLLRDPQNIPI 125
SFL C + N+++ K KR + L +D++ QKV L P IP+
Sbjct 261 SFLKCVKENDQE-KKEPKRKILGKTLTNDQIRSQKVFPLTPPCLIPV 306
Lambda K H
0.322 0.140 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1283105810
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40