bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0622_orf1
Length=95
Score E
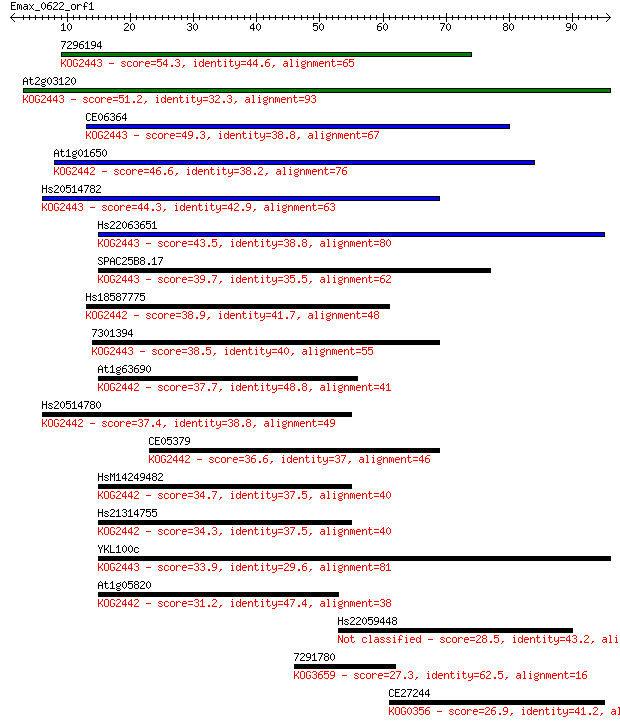
Sequences producing significant alignments: (Bits) Value
7296194 54.3 6e-08
At2g03120 51.2 5e-07
CE06364 49.3 2e-06
At1g01650 46.6 1e-05
Hs20514782 44.3 5e-05
Hs22063651 43.5 9e-05
SPAC25B8.17 39.7 0.001
Hs18587775 38.9 0.003
7301394 38.5 0.003
At1g63690 37.7 0.005
Hs20514780 37.4 0.007
CE05379 36.6 0.010
HsM14249482 34.7 0.049
Hs21314755 34.3 0.055
YKL100c 33.9 0.082
At1g05820 31.2 0.43
Hs22059448 28.5 3.4
7291780 27.3 7.0
CE27244 26.9 9.1
> 7296194
Length=389
Score = 54.3 bits (129), Expect = 6e-08, Method: Composition-based stats.
Identities = 29/65 (44%), Positives = 40/65 (61%), Gaps = 0/65 (0%)
Query 9 QRFPKFYFCVVLSFYELGLLTTGIVMLYAQHAQPALLYLVPYCLFSLFGAAALNGKIKEV 68
+R + YF L Y LGLL T VM +HAQPALLYLVP C+ + A + G++K +
Sbjct 293 KRKTRIYFYSTLIAYFLGLLATIFVMHVFKHAQPALLYLVPACMGTPLLVALIRGELKVL 352
Query 69 LAYKE 73
AY++
Sbjct 353 FAYED 357
> At2g03120
Length=344
Score = 51.2 bits (121), Expect = 5e-07, Method: Composition-based stats.
Identities = 30/93 (32%), Positives = 47/93 (50%), Gaps = 0/93 (0%)
Query 3 LAVNIHQRFPKFYFCVVLSFYELGLLTTGIVMLYAQHAQPALLYLVPYCLFSLFGAAALN 62
L ++ +R YF Y +G++ T +VM + Q AQPALLY+VP + L N
Sbjct 251 LRFDVSRRRQPQYFTSAFIGYAVGVILTIVVMNWFQAAQPALLYIVPAVIGFLASHCIWN 310
Query 63 GKIKEVLAYKEEEEAGGSTPCASAPEQAEKKEN 95
G IK +LA+ E + +T + E+ K +
Sbjct 311 GDIKPLLAFDESKTEEATTDESKTSEEVNKAHD 343
> CE06364
Length=468
Score = 49.3 bits (116), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 26/67 (38%), Positives = 35/67 (52%), Gaps = 0/67 (0%)
Query 13 KFYFCVVLSFYELGLLTTGIVMLYAQHAQPALLYLVPYCLFSLFGAAALNGKIKEVLAYK 72
++YF V + Y GL T VM + + AQPALLYLVP CLF A + G++ + Y
Sbjct 388 RYYFVVTVVAYMAGLFITMAVMHHFKAAQPALLYLVPCCLFVPLLLAVIRGELSALWNYD 447
Query 73 EEEEAGG 79
E
Sbjct 448 ESRHVDN 454
> At1g01650
Length=491
Score = 46.6 bits (109), Expect = 1e-05, Method: Composition-based stats.
Identities = 29/77 (37%), Positives = 39/77 (50%), Gaps = 6/77 (7%)
Query 8 HQRFPKFYFCVVLSFYELGLLTTGIVM-LYAQHAQPALLYLVPYCLFSLFGAAALNGKIK 66
++R YF +S Y LGLL T I + L H QPALLY+VP+ L +LF G +K
Sbjct 410 NKRLKSGYFLGTMSAYGLGLLITYIALNLMDGHGQPALLYIVPFILGTLFVLGHKRGDLK 469
Query 67 EVLAYKEEEEAGGSTPC 83
+ E + PC
Sbjct 470 TLWTTGEPDR-----PC 481
> Hs20514782
Length=385
Score = 44.3 bits (103), Expect = 5e-05, Method: Composition-based stats.
Identities = 27/64 (42%), Positives = 34/64 (53%), Gaps = 1/64 (1%)
Query 6 NIHQRFPKF-YFCVVLSFYELGLLTTGIVMLYAQHAQPALLYLVPYCLFSLFGAAALNGK 64
NI R K YF L Y +GLLT + + AQPALLYLVP+ L L A L G
Sbjct 305 NISGRMQKVSYFHCTLIGYFVGLLTATVASRIHRAAQPALLYLVPFTLLPLLTMAYLKGD 364
Query 65 IKEV 68
++ +
Sbjct 365 LRRM 368
> Hs22063651
Length=377
Score = 43.5 bits (101), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 31/87 (35%), Positives = 42/87 (48%), Gaps = 7/87 (8%)
Query 15 YFCVVLSFYELGLLTTGIVMLYAQHAQPALLYLVPYCLFSLFGAAALNGKIKEVLAYKEE 74
YF + Y GL T +M +HAQPALLYLVP C+ A G++ E+ +Y+E
Sbjct 290 YFYTSFAAYIFGLGLTIFIMHIFKHAQPALLYLVPACIGFPVLVALAKGEVTEMFSYEES 349
Query 75 EEAGGSTPC-------ASAPEQAEKKE 94
+ ASA + EKKE
Sbjct 350 NPKDPAAVTESKEGTEASASKGLEKKE 376
> SPAC25B8.17
Length=295
Score = 39.7 bits (91), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 22/62 (35%), Positives = 32/62 (51%), Gaps = 0/62 (0%)
Query 15 YFCVVLSFYELGLLTTGIVMLYAQHAQPALLYLVPYCLFSLFGAAALNGKIKEVLAYKEE 74
YF Y LGL T + Y + AQPALLYL P C+ + A ++K + +++ E
Sbjct 222 YFRNTFIAYGLGLGVTNFALYYFKAAQPALLYLSPACIVAPLLTAWYRDELKTLFSFRSE 281
Query 75 EE 76
E
Sbjct 282 TE 283
> Hs18587775
Length=684
Score = 38.9 bits (89), Expect = 0.003, Method: Composition-based stats.
Identities = 20/48 (41%), Positives = 25/48 (52%), Gaps = 0/48 (0%)
Query 13 KFYFCVVLSFYELGLLTTGIVMLYAQHAQPALLYLVPYCLFSLFGAAA 60
+ YF Y +GLL T + M+ Q QPALLYLV L + AA
Sbjct 470 QIYFVACTVAYAVGLLVTFMAMVLMQMGQPALLYLVSSTLLTSLAVAA 517
> 7301394
Length=417
Score = 38.5 bits (88), Expect = 0.003, Method: Composition-based stats.
Identities = 22/55 (40%), Positives = 33/55 (60%), Gaps = 1/55 (1%)
Query 14 FYFCVVLSFYELGLLTTGIVMLYAQHAQPALLYLVPYCLFSLFGAAALNGKIKEV 68
++ C +L ++ LGLLT + + AQPALLYLVP+ L L A L G ++ +
Sbjct 348 YFHCSLLGYF-LGLLTATVSSEVFKAAQPALLYLVPFTLLPLLLMAYLKGDLRRM 401
> At1g63690
Length=519
Score = 37.7 bits (86), Expect = 0.005, Method: Composition-based stats.
Identities = 20/42 (47%), Positives = 25/42 (59%), Gaps = 1/42 (2%)
Query 15 YFCVVLSFYELGLLTTGIVM-LYAQHAQPALLYLVPYCLFSL 55
YF + Y LGLL T + + L H QPALLY+VP+ L L
Sbjct 464 YFIWAMVAYGLGLLITYVALNLMDGHGQPALLYIVPFTLGKL 505
> Hs20514780
Length=564
Score = 37.4 bits (85), Expect = 0.007, Method: Composition-based stats.
Identities = 19/49 (38%), Positives = 26/49 (53%), Gaps = 0/49 (0%)
Query 6 NIHQRFPKFYFCVVLSFYELGLLTTGIVMLYAQHAQPALLYLVPYCLFS 54
+I + + YF Y +GLL T + + Q QPALLYLVP L +
Sbjct 408 DIQVQSSRVYFVACTIAYGVGLLVTFVALALMQRGQPALLYLVPCTLVT 456
> CE05379
Length=652
Score = 36.6 bits (83), Expect = 0.010, Method: Composition-based stats.
Identities = 17/46 (36%), Positives = 27/46 (58%), Gaps = 0/46 (0%)
Query 23 YELGLLTTGIVMLYAQHAQPALLYLVPYCLFSLFGAAALNGKIKEV 68
Y +GL+ T + + + AQPAL+YLVP LF + A G+ ++
Sbjct 549 YGIGLIVTFLALALMKTAQPALIYLVPSTLFPIIMLALCRGEFLKI 594
> HsM14249482
Length=409
Score = 34.7 bits (78), Expect = 0.049, Method: Composition-based stats.
Identities = 15/40 (37%), Positives = 24/40 (60%), Gaps = 0/40 (0%)
Query 15 YFCVVLSFYELGLLTTGIVMLYAQHAQPALLYLVPYCLFS 54
Y+ Y +G++ T +V++ + QPALLYLVP L +
Sbjct 325 YYVSSTVAYAIGMILTFVVLVLMKKGQPALLYLVPCTLIT 364
> Hs21314755
Length=520
Score = 34.3 bits (77), Expect = 0.055, Method: Composition-based stats.
Identities = 15/40 (37%), Positives = 24/40 (60%), Gaps = 0/40 (0%)
Query 15 YFCVVLSFYELGLLTTGIVMLYAQHAQPALLYLVPYCLFS 54
Y+ Y +G++ T +V++ + QPALLYLVP L +
Sbjct 436 YYVSSTVAYAIGMILTFVVLVLMKKGQPALLYLVPCTLIT 475
> YKL100c
Length=587
Score = 33.9 bits (76), Expect = 0.082, Method: Composition-based stats.
Identities = 24/81 (29%), Positives = 35/81 (43%), Gaps = 4/81 (4%)
Query 15 YFCVVLSFYELGLLTTGIVMLYAQHAQPALLYLVPYCLFSLFGAAALNGKIKEVLAYKEE 74
YF + Y L++ + + AQPALLY+VP L S A N K+ ++ +
Sbjct 450 YFITAMVSYVASLVSAMVSLSIFNTAQPALLYIVPSLLISTILVACWNKDFKQFWNFQYD 509
Query 75 EEAGGSTPCASAPEQAEKKEN 95
S + EKKEN
Sbjct 510 TIEVDK----SLKKAIEKKEN 526
> At1g05820
Length=441
Score = 31.2 bits (69), Expect = 0.43, Method: Composition-based stats.
Identities = 18/39 (46%), Positives = 23/39 (58%), Gaps = 1/39 (2%)
Query 15 YFCVVLSFYELGLLTTGI-VMLYAQHAQPALLYLVPYCL 52
YF ++ Y LGL T + + + H QPALLYLVP L
Sbjct 368 YFPWLMFGYGLGLFLTYLGLYVMNGHGQPALLYLVPCTL 406
> Hs22059448
Length=436
Score = 28.5 bits (62), Expect = 3.4, Method: Composition-based stats.
Identities = 16/38 (42%), Positives = 21/38 (55%), Gaps = 1/38 (2%)
Query 53 FSLFGAAALNGKIKEVLAYKEEEEAGG-STPCASAPEQ 89
FS G L + +V +E EEA G S PC SAP++
Sbjct 185 FSATGFWVLGFQTGQVTYCEEREEASGRSVPCLSAPQR 222
> 7291780
Length=622
Score = 27.3 bits (59), Expect = 7.0, Method: Composition-based stats.
Identities = 10/16 (62%), Positives = 12/16 (75%), Gaps = 0/16 (0%)
Query 46 YLVPYCLFSLFGAAAL 61
+LVPYCLF +FG L
Sbjct 112 FLVPYCLFLIFGGLPL 127
> CE27244
Length=568
Score = 26.9 bits (58), Expect = 9.1, Method: Composition-based stats.
Identities = 14/34 (41%), Positives = 19/34 (55%), Gaps = 0/34 (0%)
Query 61 LNGKIKEVLAYKEEEEAGGSTPCASAPEQAEKKE 94
L K+ + +A K EEAG T CA+ +A KE
Sbjct 84 LGAKLIQDVANKANEEAGDGTTCATVLARAIAKE 117
Lambda K H
0.321 0.137 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1161385214
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40