bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
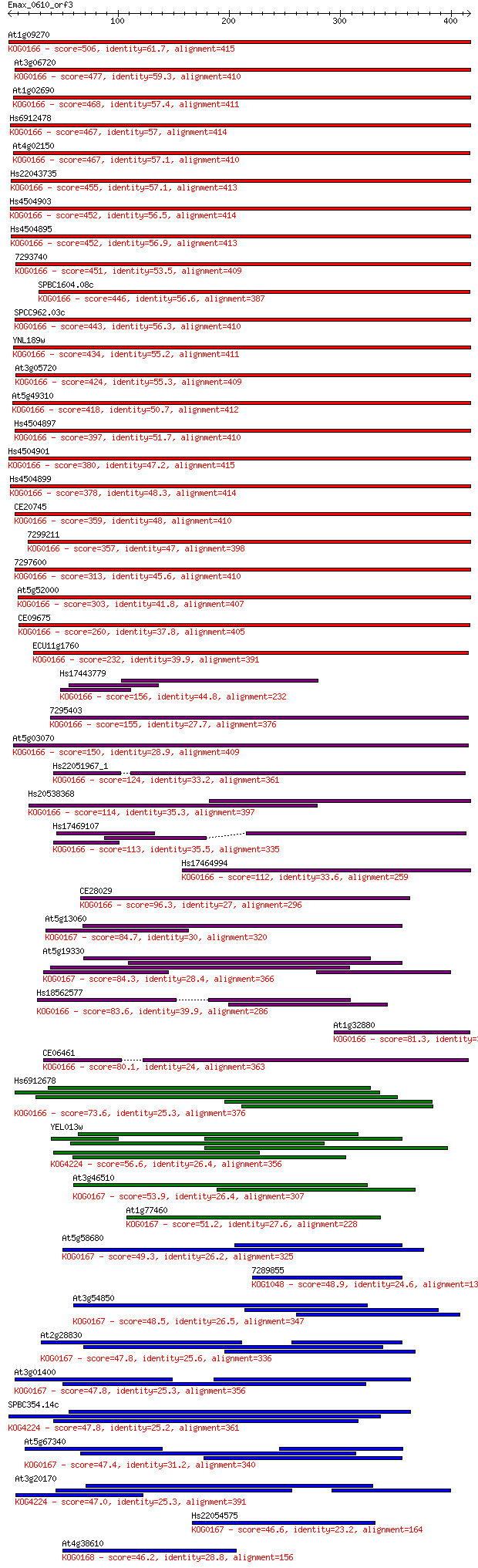
Query= Emax_0610_orf3
Length=416
Score E
Sequences producing significant alignments: (Bits) Value
At1g09270 506 4e-143
At3g06720 477 2e-134
At1g02690 468 1e-131
Hs6912478 467 2e-131
At4g02150 467 2e-131
Hs22043735 455 8e-128
Hs4504903 452 5e-127
Hs4504895 452 6e-127
7293740 451 2e-126
SPBC1604.08c 446 4e-125
SPCC962.03c 443 3e-124
YNL189w 434 2e-121
At3g05720 424 1e-118
At5g49310 418 1e-116
Hs4504897 397 2e-110
Hs4504901 380 4e-105
Hs4504899 378 1e-104
CE20745 359 7e-99
7299211 357 4e-98
7297600 313 6e-85
At5g52000 303 5e-82
CE09675 260 4e-69
ECU11g1760 232 1e-60
Hs17443779 156 8e-38
7295403 155 2e-37
At5g03070 150 5e-36
Hs22051967_1 124 6e-28
Hs20538368 114 4e-25
Hs17469107 113 9e-25
Hs17464994 112 1e-24
CE28029 96.3 1e-19
At5g13060 84.7 4e-16
At5g19330 84.3 5e-16
Hs18562577 83.6 7e-16
At1g32880 81.3 4e-15
CE06461 80.1 9e-15
Hs6912678 73.6 7e-13
YEL013w 56.6 1e-07
At3g46510 53.9 7e-07
At1g77460 51.2 4e-06
At5g58680 49.3 2e-05
7289855 48.9 2e-05
At3g54850 48.5 3e-05
At2g28830 47.8 5e-05
At3g01400 47.8 5e-05
SPBC354.14c 47.8 5e-05
At5g67340 47.4 5e-05
At3g20170 47.0 7e-05
Hs22054575 46.6 9e-05
At4g38610 46.2 1e-04
> At1g09270
Length=538
Score = 506 bits (1302), Expect = 4e-143, Method: Compositional matrix adjust.
Identities = 256/418 (61%), Positives = 313/418 (74%), Gaps = 7/418 (1%)
Query 2 LEGVDLQQLVQALASGVPDAELEGTQLFRRALSLENNPPIQEVIDAGAVPHFVRFLSMHG 61
LEG+ + +VQ + S P A+LE T FR+ LS+E +PPI EVI AG +P FV FL H
Sbjct 78 LEGIPM--MVQGVYSDDPQAQLEATTQFRKLLSIERSPPIDEVIKAGVIPRFVEFLGRHD 135
Query 62 RYQLQFEAAWALTNIASGTQEQTRVVVECGAIPLFVELLRCPKEEVREQAVWALGNIAGD 121
QLQFEAAWALTN+ASGT + TRVV+E GA+P+FV+LL ++VREQAVWALGN+AGD
Sbjct 136 HPQLQFEAAWALTNVASGTSDHTRVVIEQGAVPIFVKLLTSASDDVREQAVWALGNVAGD 195
Query 122 SPSCRNLVLDGGALPPLLEQLNTPGSKVAMLRNATWTLSNLCRGKPAPVFEKVSPALPAL 181
SP+CRNLVL+ GAL PLL QLN SK++MLRNATWTLSN CRGKP FE+V PALP L
Sbjct 196 SPNCRNLVLNYGALEPLLAQLNE-NSKLSMLRNATWTLSNFCRGKPPTPFEQVKPALPIL 254
Query 182 AVLIYSDDAEVLTDACWALSYLSDGPNDRIKAVLEAGVCARLVELLSHQSNLVQTPALRA 241
LIY +D EVLTDACWALSYLSDGPND+I+AV+EAGVC RLVELL HQS V PALR
Sbjct 255 RQLIYLNDEEVLTDACWALSYLSDGPNDKIQAVIEAGVCPRLVELLGHQSPTVLIPALRT 314
Query 242 VGNIVTGDDTQTQVVLTCGALPKLLTLLSST-KKNVQKEACWTVSNITAGNKDQIQEVLD 300
VGNIVTGDD+QTQ ++ G LP L LL+ KK+++KEACWT+SNITAGNK QI+ V+
Sbjct 315 VGNIVTGDDSQTQFIIESGVLPHLYNLLTQNHKKSIKKEACWTISNITAGNKLQIEAVVG 374
Query 301 SGAIPALLHLLETADFDTKKEAAWAVSNAVSGGTDMQVALLVEKGCIPPLCSLLGCMDNK 360
+G I L+HLL+ A+FD KKEAAWA+SNA SGG+ Q+ LV +GCI PLC LL C D +
Sbjct 375 AGIILPLVHLLQNAEFDIKKEAAWAISNATSGGSHEQIQYLVTQGCIKPLCDLLICPDPR 434
Query 361 IIMVALEALNAILRTGKKIKETEGLS--VNPYATLIEQADGATALEKLQDSTNENVFK 416
I+ V LE L IL+ G+ KE GL+ VN YA +IE++DG +E LQ N +++
Sbjct 435 IVTVCLEGLENILKVGEADKEM-GLNSGVNLYAQIIEESDGLDKVENLQSHDNNEIYE 491
> At3g06720
Length=532
Score = 477 bits (1228), Expect = 2e-134, Method: Compositional matrix adjust.
Identities = 243/412 (58%), Positives = 300/412 (72%), Gaps = 3/412 (0%)
Query 7 LQQLVQALASGVPDAELEGTQLFRRALSLENNPPIQEVIDAGAVPHFVRFLSMHGRYQLQ 66
L+ +V + S P +LE T FR+ LS+E +PPI+EVI AG VP FV FL +Q
Sbjct 74 LKDMVAGVWSDDPALQLESTTQFRKLLSIERSPPIEEVISAGVVPRFVEFLKKEDYPAIQ 133
Query 67 FEAAWALTNIASGTQEQTRVVVECGAIPLFVELLRCPKEEVREQAVWALGNIAGDSPSCR 126
FEAAWALTNIASGT + T+VV++ A+P+FV+LL P ++VREQAVWALGN+AGDSP CR
Sbjct 134 FEAAWALTNIASGTSDHTKVVIDHNAVPIFVQLLASPSDDVREQAVWALGNVAGDSPRCR 193
Query 127 NLVLDGGALPPLLEQLNTPGSKVAMLRNATWTLSNLCRGKPAPVFEKVSPALPALAVLIY 186
+LVL GAL PLL QLN +K++MLRNATWTLSN CRGKP P F++V PALPAL LI+
Sbjct 194 DLVLGCGALLPLLNQLNE-HAKLSMLRNATWTLSNFCRGKPQPHFDQVKPALPALERLIH 252
Query 187 SDDAEVLTDACWALSYLSDGPNDRIKAVLEAGVCARLVELLSHQSNLVQTPALRAVGNIV 246
SDD EVLTDACWALSYLSDG ND+I+ V++AGV +LVELL H S V PALR VGNIV
Sbjct 253 SDDEEVLTDACWALSYLSDGTNDKIQTVIQAGVVPKLVELLLHHSPSVLIPALRTVGNIV 312
Query 247 TGDDTQTQVVLTCGALPKLLTLLSST-KKNVQKEACWTVSNITAGNKDQIQEVLDSGAIP 305
TGDD QTQ V+ GALP L LL+ KK+++KEACWT+SNITAGNKDQIQ V+++ I
Sbjct 313 TGDDIQTQCVINSGALPCLANLLTQNHKKSIKKEACWTISNITAGNKDQIQTVVEANLIS 372
Query 306 ALLHLLETADFDTKKEAAWAVSNAVSGGTDMQVALLVEKGCIPPLCSLLGCMDNKIIMVA 365
L+ LL+ A+FD KKEAAWA+SNA SGG+ Q+ LVE+GCI PLC LL C D +II V
Sbjct 373 PLVSLLQNAEFDIKKEAAWAISNATSGGSHDQIKYLVEQGCIKPLCDLLVCPDPRIITVC 432
Query 366 LEALNAILRTGKKIKE-TEGLSVNPYATLIEQADGATALEKLQDSTNENVFK 416
LE L IL+ G+ K +N YA LI+ A+G +E LQ N +++
Sbjct 433 LEGLENILKVGEAEKNLGHTGDMNYYAQLIDDAEGLEKIENLQSHDNNEIYE 484
> At1g02690
Length=538
Score = 468 bits (1204), Expect = 1e-131, Method: Compositional matrix adjust.
Identities = 236/413 (57%), Positives = 294/413 (71%), Gaps = 3/413 (0%)
Query 6 DLQQLVQALASGVPDAELEGTQLFRRALSLENNPPIQEVIDAGAVPHFVRFLSMHGRYQL 65
++QQ++ + S D +LE T FRR LS+E NPPI EV+ +G VPH V+FLS QL
Sbjct 77 NIQQMIAGVMSEDRDLQLEATASFRRLLSIERNPPINEVVQSGVVPHIVQFLSRDDFTQL 136
Query 66 QFEAAWALTNIASGTQEQTRVVVECGAIPLFVELLRCPKEEVREQAVWALGNIAGDSPSC 125
QFEAAWALTNIASGT E TRV+++ GA+PLFV+LL EEVREQAVWALGN+AGDSP C
Sbjct 137 QFEAAWALTNIASGTSENTRVIIDSGAVPLFVKLLSSASEEVREQAVWALGNVAGDSPKC 196
Query 126 RNLVLDGGALPPLLEQLNTPGSKVAMLRNATWTLSNLCRGKPAPVFEKVSPALPALAVLI 185
R+ VL A+ LL Q + SK++MLRNATWTLSN CRGKP P FE+ ALPAL L+
Sbjct 197 RDHVLSCEAMMSLLAQFHE-HSKLSMLRNATWTLSNFCRGKPQPAFEQTKAALPALERLL 255
Query 186 YSDDAEVLTDACWALSYLSDGPNDRIKAVLEAGVCARLVELLSHQSNLVQTPALRAVGNI 245
+S D EVLTDA WALSYLSDG N++I+ V++AGV RLV+LL+H S V PALR +GNI
Sbjct 256 HSTDEEVLTDASWALSYLSDGTNEKIQTVIDAGVIPRLVQLLAHPSPSVLIPALRTIGNI 315
Query 246 VTGDDTQTQVVLTCGALPKLLTLLSST-KKNVQKEACWTVSNITAGNKDQIQEVLDSGAI 304
VTGDD QTQ V++ ALP LL LL +T KK+++KEACWT+SNITAGN QIQEV +G I
Sbjct 316 VTGDDIQTQAVISSQALPGLLNLLKNTYKKSIKKEACWTISNITAGNTSQIQEVFQAGII 375
Query 305 PALLHLLETADFDTKKEAAWAVSNAVSGGTDMQVALLVEKGCIPPLCSLLGCMDNKIIMV 364
L++LLE +F+ KKEA WA+SNA SGG Q+ LV +GCI PLC LL C D +++ V
Sbjct 376 RPLINLLEIGEFEIKKEAVWAISNATSGGNHDQIKFLVSQGCIRPLCDLLPCPDPRVVTV 435
Query 365 ALEALNAILRTGKKIKETEGL-SVNPYATLIEQADGATALEKLQDSTNENVFK 416
LE L IL+ G+ K + N YA +IE ADG +E LQ N +++
Sbjct 436 TLEGLENILKVGEAEKNLGNTGNDNLYAQMIEDADGLDKIENLQSHDNNEIYE 488
> Hs6912478
Length=536
Score = 467 bits (1201), Expect = 2e-131, Method: Compositional matrix adjust.
Identities = 236/416 (56%), Positives = 300/416 (72%), Gaps = 3/416 (0%)
Query 3 EGVDLQQLVQALASGVPDAELEGTQLFRRALSLENNPPIQEVIDA-GAVPHFVRFLSMHG 61
E V +++V+ L S D +L TQ FR+ LS E +PPI EVI+ V FV FL +
Sbjct 78 ESVITREMVEMLFSDDSDLQLATTQKFRKLLSKEPSPPIDEVINTPRVVDRFVEFLKRNE 137
Query 62 RYQLQFEAAWALTNIASGTQEQTRVVVECGAIPLFVELLRCPKEEVREQAVWALGNIAGD 121
LQFEAAWALTNIASGT +QT++V+E GA+P+F+ELL E+V+EQAVWALGNIAGD
Sbjct 138 NCTLQFEAAWALTNIASGTSQQTKIVIEAGAVPIFIELLNSDFEDVQEQAVWALGNIAGD 197
Query 122 SPSCRNLVLDGGALPPLLEQLNTPGSKVAMLRNATWTLSNLCRGK-PAPVFEKVSPALPA 180
S CR+ VL+ L PLL L T +++ M RNA W LSNLCRGK P P F KVSP LP
Sbjct 198 SSVCRDYVLNCSILNPLLTLL-TKSTRLTMTRNAVWALSNLCRGKNPPPEFAKVSPCLPV 256
Query 181 LAVLIYSDDAEVLTDACWALSYLSDGPNDRIKAVLEAGVCARLVELLSHQSNLVQTPALR 240
L+ L++S D+++L DACWALSYLSDGPN++I+AV+++GVC RLVELL H V +PALR
Sbjct 257 LSRLLFSSDSDLLADACWALSYLSDGPNEKIQAVIDSGVCRRLVELLMHNDYKVASPALR 316
Query 241 AVGNIVTGDDTQTQVVLTCGALPKLLTLLSSTKKNVQKEACWTVSNITAGNKDQIQEVLD 300
AVGNIVTGDD QTQV+L C ALP LL LLSS K++++KEACWT+SNITAGN+ QIQ V+D
Sbjct 317 AVGNIVTGDDIQTQVILNCSALPCLLHLLSSPKESIRKEACWTISNITAGNRAQIQAVID 376
Query 301 SGAIPALLHLLETADFDTKKEAAWAVSNAVSGGTDMQVALLVEKGCIPPLCSLLGCMDNK 360
+ P L+ +L+ A+F T+KEAAWA++NA SGGT Q+ LV GCI PLC LL MD+K
Sbjct 377 ANIFPVLIEILQKAEFRTRKEAAWAITNATSGGTPEQIRYLVSLGCIKPLCDLLTVMDSK 436
Query 361 IIMVALEALNAILRTGKKIKETEGLSVNPYATLIEQADGATALEKLQDSTNENVFK 416
I+ VAL L ILR G++ + G VNPY LIE+A G +E LQ N+ +++
Sbjct 437 IVQVALNGLENILRLGEQEGKRSGSGVNPYCGLIEEAYGLDKIEFLQSHENQEIYQ 492
> At4g02150
Length=531
Score = 467 bits (1201), Expect = 2e-131, Method: Compositional matrix adjust.
Identities = 234/412 (56%), Positives = 297/412 (72%), Gaps = 3/412 (0%)
Query 6 DLQQLVQALASGVPDAELEGTQLFRRALSLENNPPIQEVIDAGAVPHFVRFLSMHGRYQL 65
+L +V + S +++LE T L R+ LS+E NPPI EV+ +G VP V+FLS +L
Sbjct 76 NLPAMVAGIWSEDSNSQLEATNLLRKLLSIEQNPPINEVVQSGVVPRVVKFLSRDDFPKL 135
Query 66 QFEAAWALTNIASGTQEQTRVVVECGAIPLFVELLRCPKEEVREQAVWALGNIAGDSPSC 125
QFEAAWALTNIASGT E T V++E GA+P+F++LL E+VREQAVWALGN+AGDSP C
Sbjct 136 QFEAAWALTNIASGTSENTNVIIESGAVPIFIQLLSSASEDVREQAVWALGNVAGDSPKC 195
Query 126 RNLVLDGGALPPLLEQLNTPGSKVAMLRNATWTLSNLCRGKPAPVFEKVSPALPALAVLI 185
R+LVL GA+ PLL Q N +K++MLRNATWTLSN CRGKP P FE+ PALP L L+
Sbjct 196 RDLVLSYGAMTPLLSQFNE-NTKLSMLRNATWTLSNFCRGKPPPAFEQTQPALPVLERLV 254
Query 186 YSDDAEVLTDACWALSYLSDGPNDRIKAVLEAGVCARLVELLSHQSNLVQTPALRAVGNI 245
S D EVLTDACWALSYLSD ND+I+AV+EAGV RL++LL H S V PALR +GNI
Sbjct 255 QSMDEEVLTDACWALSYLSDNSNDKIQAVIEAGVVPRLIQLLGHSSPSVLIPALRTIGNI 314
Query 246 VTGDDTQTQVVLTCGALPKLLTLLSST-KKNVQKEACWTVSNITAGNKDQIQEVLDSGAI 304
VTGDD QTQ+VL ALP LL LL + KK+++KEACWT+SNITAGN DQIQ V+D+G I
Sbjct 315 VTGDDLQTQMVLDQQALPCLLNLLKNNYKKSIKKEACWTISNITAGNADQIQAVIDAGII 374
Query 305 PALLHLLETADFDTKKEAAWAVSNAVSGGTDMQVALLVEKGCIPPLCSLLGCMDNKIIMV 364
+L+ +L++A+F+ KKEAAW +SNA SGGT Q+ +V +GCI PLC LL C D K++ V
Sbjct 375 QSLVWVLQSAEFEVKKEAAWGISNATSGGTHDQIKFMVSQGCIKPLCDLLTCPDLKVVTV 434
Query 365 ALEALNAILRTGKKIKETEGLSV-NPYATLIEQADGATALEKLQDSTNENVF 415
LEAL IL G+ K N YA +I++A+G +E LQ N +++
Sbjct 435 CLEALENILVVGEAEKNLGHTGEDNLYAQMIDEAEGLEKIENLQSHDNNDIY 486
> Hs22043735
Length=538
Score = 455 bits (1170), Expect = 8e-128, Method: Compositional matrix adjust.
Identities = 236/415 (56%), Positives = 296/415 (71%), Gaps = 3/415 (0%)
Query 4 GVDLQQLVQALASGVPDAELEGTQLFRRALSLENNPPIQEVIDA-GAVPHFVRFLSMHGR 62
GV +++ + S P+ +L TQ FR+ LS E NPPI EVI G V FV FL
Sbjct 81 GVITSDMIEMIFSKSPEQQLSATQKFRKLLSKEPNPPIDEVISTPGVVARFVEFLKRKEN 140
Query 63 YQLQFEAAWALTNIASGTQEQTRVVVECGAIPLFVELLRCPKEEVREQAVWALGNIAGDS 122
LQFE+AW LTNIASG QTR+V++ GA+P+F+ELL E+V+EQAVWALGNIAGDS
Sbjct 141 CTLQFESAWVLTNIASGNSLQTRIVIQAGAVPIFIELLSSEFEDVQEQAVWALGNIAGDS 200
Query 123 PSCRNLVLDGGALPPLLEQLNTPGSKVAMLRNATWTLSNLCRGK-PAPVFEKVSPALPAL 181
CR+ VLD LPPLL QL + +++ M RNA W LSNLCRGK P P F KVSP L L
Sbjct 201 TMCRDYVLDCNILPPLL-QLFSKQNRLTMTRNAVWALSNLCRGKSPPPEFAKVSPCLNVL 259
Query 182 AVLIYSDDAEVLTDACWALSYLSDGPNDRIKAVLEAGVCARLVELLSHQSNLVQTPALRA 241
+ L++ D +VL DACWALSYLSDGPND+I+AV++AGVC RLVELL H V +PALRA
Sbjct 260 SWLLFVSDTDVLADACWALSYLSDGPNDKIQAVIDAGVCRRLVELLMHNDYKVVSPALRA 319
Query 242 VGNIVTGDDTQTQVVLTCGALPKLLTLLSSTKKNVQKEACWTVSNITAGNKDQIQEVLDS 301
VGNIVTGDD QTQV+L C AL LL LLSS K++++KEACWT+SNITAGN+ QIQ V+D+
Sbjct 320 VGNIVTGDDIQTQVILNCSALQSLLHLLSSPKESIKKEACWTISNITAGNRAQIQTVIDA 379
Query 302 GAIPALLHLLETADFDTKKEAAWAVSNAVSGGTDMQVALLVEKGCIPPLCSLLGCMDNKI 361
PAL+ +L+TA+F T+KEAAWA++NA SGG+ Q+ LVE GCI PLC LL MD+KI
Sbjct 380 NIFPALISILQTAEFRTRKEAAWAITNATSGGSAEQIKYLVELGCIKPLCDLLTVMDSKI 439
Query 362 IMVALEALNAILRTGKKIKETEGLSVNPYATLIEQADGATALEKLQDSTNENVFK 416
+ VAL L ILR G++ + G +NPY LIE+A G +E LQ N+ +++
Sbjct 440 VQVALNGLENILRLGEQEAKRNGTGINPYCALIEEAYGLDKIEFLQSHENQEIYQ 494
> Hs4504903
Length=536
Score = 452 bits (1164), Expect = 5e-127, Method: Compositional matrix adjust.
Identities = 234/416 (56%), Positives = 295/416 (70%), Gaps = 3/416 (0%)
Query 3 EGVDLQQLVQALASGVPDAELEGTQLFRRALSLENNPPIQEVIDA-GAVPHFVRFLSMHG 61
EGV +VQ + S D +L TQ FR+ LS E NPPI +VI G V FV+FL +
Sbjct 78 EGVVTTDMVQMIFSNNADQQLTATQKFRKLLSKEPNPPIDQVIQKPGVVQRFVKFLERNE 137
Query 62 RYQLQFEAAWALTNIASGTQEQTRVVVECGAIPLFVELLRCPKEEVREQAVWALGNIAGD 121
LQFEAAWALTNIASGT T+VV+E GA+P+F++LL E+V+EQAVWALGNIAGD
Sbjct 138 NCTLQFEAAWALTNIASGTFLHTKVVIETGAVPIFIKLLNSEHEDVQEQAVWALGNIAGD 197
Query 122 SPSCRNLVLDGGALPPLLEQLNTPGSKVAMLRNATWTLSNLCRGK-PAPVFEKVSPALPA 180
+ CR+ VL+ LP L +L T +++ RNA W LSNLCRGK P P F KVSP L
Sbjct 198 NAECRDFVLNCEILP-PLLELLTNSNRLTTTRNAVWALSNLCRGKNPPPNFSKVSPCLNV 256
Query 181 LAVLIYSDDAEVLTDACWALSYLSDGPNDRIKAVLEAGVCARLVELLSHQSNLVQTPALR 240
L+ L++S D +VL D CWALSYLSDGPND+I+AV+++GVC RLVELL H V +PALR
Sbjct 257 LSRLLFSSDPDVLADVCWALSYLSDGPNDKIQAVIDSGVCRRLVELLMHNDYKVVSPALR 316
Query 241 AVGNIVTGDDTQTQVVLTCGALPKLLTLLSSTKKNVQKEACWTVSNITAGNKDQIQEVLD 300
AVGNIVTGDD QTQV+L C ALP LL LLSS K++++KEACWTVSNITAGN+ QIQ V+D
Sbjct 317 AVGNIVTGDDIQTQVILNCSALPCLLHLLSSPKESIRKEACWTVSNITAGNRAQIQAVID 376
Query 301 SGAIPALLHLLETADFDTKKEAAWAVSNAVSGGTDMQVALLVEKGCIPPLCSLLGCMDNK 360
+ P L+ +L+ A+F T+KEAAWA++NA SGGT Q+ LV GCI PLC LL MD+K
Sbjct 377 ANIFPVLIEILQKAEFRTRKEAAWAITNATSGGTPEQIRYLVALGCIKPLCDLLTVMDSK 436
Query 361 IIMVALEALNAILRTGKKIKETEGLSVNPYATLIEQADGATALEKLQDSTNENVFK 416
I+ VAL L ILR G++ + G+ +NPY LIE+A G +E LQ N+ +++
Sbjct 437 IVQVALNGLENILRLGEQESKQNGIGINPYCALIEEAYGLDKIEFLQSHENQEIYQ 492
> Hs4504895
Length=538
Score = 452 bits (1163), Expect = 6e-127, Method: Compositional matrix adjust.
Identities = 235/415 (56%), Positives = 295/415 (71%), Gaps = 3/415 (0%)
Query 4 GVDLQQLVQALASGVPDAELEGTQLFRRALSLENNPPIQEVIDA-GAVPHFVRFLSMHGR 62
GV +++ + S P+ +L TQ FR+ LS E NPPI EVI G V FV FL
Sbjct 81 GVITSDMIEMIFSKSPEQQLSATQKFRKLLSKEPNPPIDEVISTPGVVARFVEFLKRKEN 140
Query 63 YQLQFEAAWALTNIASGTQEQTRVVVECGAIPLFVELLRCPKEEVREQAVWALGNIAGDS 122
LQFE+AW LTNIASG QTR+V++ A+P+F+ELL E+V+EQAVWALGNIAGDS
Sbjct 141 CSLQFESAWVLTNIASGNSLQTRIVIQARAVPIFIELLSSEFEDVQEQAVWALGNIAGDS 200
Query 123 PSCRNLVLDGGALPPLLEQLNTPGSKVAMLRNATWTLSNLCRGK-PAPVFEKVSPALPAL 181
CR+ VLD LPPLL QL + +++ M RNA W LSNLCRGK P P F KVSP L L
Sbjct 201 TMCRDYVLDCNILPPLL-QLFSKQNRLTMTRNAVWALSNLCRGKSPPPEFAKVSPCLNVL 259
Query 182 AVLIYSDDAEVLTDACWALSYLSDGPNDRIKAVLEAGVCARLVELLSHQSNLVQTPALRA 241
+ L++ D +VL DACWALSYLSDGPND+I+AV++AGVC RLVELL H V +PALRA
Sbjct 260 SWLLFVSDTDVLADACWALSYLSDGPNDKIQAVIDAGVCRRLVELLMHNDYKVVSPALRA 319
Query 242 VGNIVTGDDTQTQVVLTCGALPKLLTLLSSTKKNVQKEACWTVSNITAGNKDQIQEVLDS 301
VGNIVTGDD QTQV+L C AL LL LLSS K++++KEACWT+SNITAGN+ QIQ V+D+
Sbjct 320 VGNIVTGDDIQTQVILNCSALQSLLHLLSSPKESIKKEACWTISNITAGNRAQIQTVIDA 379
Query 302 GAIPALLHLLETADFDTKKEAAWAVSNAVSGGTDMQVALLVEKGCIPPLCSLLGCMDNKI 361
PAL+ +L+TA+F T+KEAAWA++NA SGG+ Q+ LVE GCI PLC LL MD+KI
Sbjct 380 NIFPALISILQTAEFRTRKEAAWAITNATSGGSAEQIKYLVELGCIKPLCDLLTVMDSKI 439
Query 362 IMVALEALNAILRTGKKIKETEGLSVNPYATLIEQADGATALEKLQDSTNENVFK 416
+ VAL L ILR G++ + G +NPY LIE+A G +E LQ N+ +++
Sbjct 440 VQVALNGLENILRLGEQEAKRNGTGINPYCALIEEAYGLDKIEFLQSHENQEIYQ 494
> 7293740
Length=543
Score = 451 bits (1159), Expect = 2e-126, Method: Compositional matrix adjust.
Identities = 219/410 (53%), Positives = 289/410 (70%), Gaps = 6/410 (1%)
Query 8 QQLVQALASGVPDAELEGTQLFRRALSLENNPPIQEVIDAGAVPHFVRFLSMHGRYQLQF 67
++++Q L SG + +LE TQ FR+ LS + NPPI+EVI G VP FV FL LQF
Sbjct 97 EEMIQMLFSGRENEQLEATQKFRKLLSRDPNPPIEEVIQKGIVPQFVTFLRNSANATLQF 156
Query 68 EAAWALTNIASGTQEQTRVVVECGAIPLFVELLRCPKEEVREQAVWALGNIAGDSPSCRN 127
EAAW LTNIASGT +QT+VV+E GA+P+F++LL P ++V+EQAVWALGNIAGDSP CR+
Sbjct 157 EAAWTLTNIASGTSQQTKVVIEAGAVPIFIDLLSSPHDDVQEQAVWALGNIAGDSPMCRD 216
Query 128 LVLDGGALPPLLEQLNTPGSKVAMLRNATWTLSNLCRGKPAPV-FEKVSPALPALAVLIY 186
+L G L PLL L+ ++ M+RNA WTLSNLCRGK P F K+S LP LA L+
Sbjct 217 HLLGSGILEPLLHVLSN-SDRITMIRNAVWTLSNLCRGKSPPADFAKISHGLPILARLLK 275
Query 187 SDDAEVLTDACWALSYLSDGPNDRIKAVLEAGVCARLVELLSHQSNLVQTPALRAVGNIV 246
DA+VL+D CWA+ YLSDGPND+I+AV++AGVC RLVELL H V T ALRAVGNIV
Sbjct 276 YTDADVLSDTCWAIGYLSDGPNDKIQAVIDAGVCRRLVELLLHPQQNVSTAALRAVGNIV 335
Query 247 TGDDTQTQVVLTCGALPKLLTLLSSTKKNVQKEACWTVSNITAGNKDQIQEVLDSGAIPA 306
TGDD QTQV+L AL + LL ST + ++KE+CWT+SNI AGN++QIQ ++++ P
Sbjct 336 TGDDQQTQVILGYNALTCISHLLHSTAETIKKESCWTISNIAAGNREQIQALINANIFPQ 395
Query 307 LLHLLETADFDTKKEAAWAVSNAVSGGTDMQVALLVEKGCIPPLCSLLGCMDNKIIMVAL 366
L+ +++TA+F T+KEAAWA++NA S GT Q+ LV+ GC+PP+C L +D+ I+ VAL
Sbjct 396 LMVIMQTAEFKTRKEAAWAITNATSSGTHEQIHYLVQVGCVPPMCDFLTVVDSDIVQVAL 455
Query 367 EALNAILRTGKKIKETEGLSVNPYATLIEQADGATALEKLQDSTNENVFK 416
AL IL+ G+K + NPYA IE+ G +E LQ N +++
Sbjct 456 NALENILKAGEKFQTRP----NPYAITIEECGGLDKIEYLQAHENRDIYH 501
> SPBC1604.08c
Length=539
Score = 446 bits (1147), Expect = 4e-125, Method: Compositional matrix adjust.
Identities = 219/390 (56%), Positives = 282/390 (72%), Gaps = 4/390 (1%)
Query 29 FRRALSLENNPPIQEVIDAGAVPHFVRFLSMHGRYQLQFEAAWALTNIASGTQEQTRVVV 88
FR+ LS E +PPI +VI G V FV+FL + LQFEAAWALTNIASGT +QTR+VV
Sbjct 101 FRKYLSKETHPPIDQVIACGVVDRFVQFLESE-HHLLQFEAAWALTNIASGTTDQTRIVV 159
Query 89 ECGAIPLFVELLRCPKEEVREQAVWALGNIAGDSPSCRNLVLDGGALPPLLEQLNTPGSK 148
+ GA+P F++LL P+++VREQ VWALGNIAGDS +CR+ VL G L PLL L + S
Sbjct 160 DSGAVPRFIQLLSSPEKDVREQVVWALGNIAGDSSACRDYVLGNGVLQPLLNILQSSASD 219
Query 149 VAMLRNATWTLSNLCRGK-PAPVFEKVSPALPALAVLIYSDDAEVLTDACWALSYLSDGP 207
V+MLRNATWTLSNLCRGK P P + +S A+P LA L+YS+D E++ DACWA+SYLSDGP
Sbjct 220 VSMLRNATWTLSNLCRGKNPPPNWSTISVAVPILAKLLYSEDVEIIVDACWAISYLSDGP 279
Query 208 NDRIKAVLEAGVCARLVELLSHQSNLVQTPALRAVGNIVTGDDTQTQVVLTCGALPKLLT 267
N++I A+L+ G RLVELLS S +QTPALR+VGNIVTG D QTQ+++ CGAL +
Sbjct 280 NEKIGAILDVGCAPRLVELLSSPSVNIQTPALRSVGNIVTGTDAQTQIIIDCGALNAFPS 339
Query 268 LLSSTKKNVQKEACWTVSNITAGNKDQIQEVLDSGAIPALLHLLETADFDTKKEAAWAVS 327
LLS K+N++KEACWT+SNITAGN QIQ +++S IP L+HLL AD+ TKKEA WA+S
Sbjct 340 LLSHQKENIRKEACWTISNITAGNTQQIQAIIESNLIPPLVHLLSYADYKTKKEACWAIS 399
Query 328 NAVSGGTDM--QVALLVEKGCIPPLCSLLGCMDNKIIMVALEALNAILRTGKKIKETEGL 385
NA SGG Q+ LV +G I PLC +L DNKII VAL+A+ IL+ G+ + +
Sbjct 400 NATSGGLGQPDQIRYLVSQGVIKPLCDMLNGSDNKIIQVALDAIENILKVGEMDRTMDLE 459
Query 386 SVNPYATLIEQADGATALEKLQDSTNENVF 415
++N YA +E+A G + LQ S N +++
Sbjct 460 NINQYAVYVEEAGGMDMIHDLQSSGNNDIY 489
> SPCC962.03c
Length=542
Score = 443 bits (1140), Expect = 3e-124, Method: Compositional matrix adjust.
Identities = 231/413 (55%), Positives = 299/413 (72%), Gaps = 5/413 (1%)
Query 7 LQQLVQALASGVPDAELEGTQLFRRALSLENNPPIQEVIDAGAVPHFVRFLSMHGRYQLQ 66
L L++AL S +A+++ T FR+ALS E NPPIQ+VIDAG VP FV FLS H L+
Sbjct 80 LPDLLKALYSDDIEAQIQATAKFRKALSKETNPPIQKVIDAGVVPRFVEFLS-HENNLLK 138
Query 67 FEAAWALTNIASGTQEQTRVVVECGAIPLFVELLRCPKEEVREQAVWALGNIAGDSPSCR 126
FEA+WALTN+ASG+ QT VVVE A+P+FV LL +++VREQAVWALGNIAGDSP CR
Sbjct 139 FEASWALTNVASGSSNQTHVVVEANAVPVFVSLLSSSEQDVREQAVWALGNIAGDSPMCR 198
Query 127 NLVLDGGALPPLLEQLNTPGSKVAMLRNATWTLSNLCRGK-PAPVFEKVSPALPALAVLI 185
+ VL G L PLL + + +++MLRN+TWTLSN+CRGK P P + +S +P L+ LI
Sbjct 199 DHVLQCGVLEPLLNIIES-NRRLSMLRNSTWTLSNMCRGKNPQPDWNSISQVIPVLSKLI 257
Query 186 YSDDAEVLTDACWALSYLSDGPNDRIKAVLEAGVCARLVELLSHQSNLVQTPALRAVGNI 245
Y+ D +VL DA WA+SYLSDG N++I+A+++AG+ RLVELL H S VQTPALR+VGNI
Sbjct 258 YTLDEDVLVDALWAISYLSDGANEKIQAIIDAGIPRRLVELLMHPSAQVQTPALRSVGNI 317
Query 246 VTGDDTQTQVVLTCGALPKLLTLLSSTKKNVQKEACWTVSNITAGNKDQIQEVLDSGAIP 305
VTGDD QTQV++ CGAL LL+LLSS + V+KEACWT+SNITAGN QIQ V+++ IP
Sbjct 318 VTGDDVQTQVIINCGALSALLSLLSSPRDGVRKEACWTISNITAGNSSQIQYVIEANIIP 377
Query 306 ALLHLLETADFDTKKEAAWAVSNAVSGGTDM--QVALLVEKGCIPPLCSLLGCMDNKIIM 363
L+HLL TADF +KEA WA+SNA SGG Q+ LVE+G I PLC+LL C DNKII
Sbjct 378 PLIHLLTTADFKIQKEACWAISNATSGGARRPDQIRYLVEQGAIKPLCNLLACQDNKIIQ 437
Query 364 VALEALNAILRTGKKIKETEGLSVNPYATLIEQADGATALEKLQDSTNENVFK 416
VAL+ + ILR G+ + +N YA +E A G + + Q+S+N +++
Sbjct 438 VALDGIENILRVGELDRANNPDKINLYAVYVEDAGGMDLIHECQNSSNSEIYQ 490
> YNL189w
Length=542
Score = 434 bits (1115), Expect = 2e-121, Method: Compositional matrix adjust.
Identities = 227/414 (54%), Positives = 286/414 (69%), Gaps = 5/414 (1%)
Query 6 DLQQLVQALASGVPDAELEGTQLFRRALSLENNPPIQEVIDAGAVPHFVRFLSMHGRYQL 65
+L Q+ Q L S +L T FR+ LS E+ PPI VI AG VP V F+ + L
Sbjct 88 ELPQMTQQLNSDDMQEQLSATVKFRQILSREHRPPIDVVIQAGVVPRLVEFMRENQPEML 147
Query 66 QFEAAWALTNIASGTQEQTRVVVECGAIPLFVELLRCPKEEVREQAVWALGNIAGDSPSC 125
Q EAAWALTNIASGT QT+VVV+ A+PLF++LL EV+EQA+WALGN+AGDS
Sbjct 148 QLEAAWALTNIASGTSAQTKVVVDADAVPLFIQLLYTGSVEVKEQAIWALGNVAGDSTDY 207
Query 126 RNLVLDGGALPPLLEQLNTPGSKVAMLRNATWTLSNLCRGK-PAPVFEKVSPALPALAVL 184
R+ VL A+ P+L N+ +K +++R ATWTLSNLCRGK P P + VS ALP LA L
Sbjct 208 RDYVLQCNAMEPILGLFNS--NKPSLIRTATWTLSNLCRGKKPQPDWSVVSQALPTLAKL 265
Query 185 IYSDDAEVLTDACWALSYLSDGPNDRIKAVLEAGVCARLVELLSHQSNLVQTPALRAVGN 244
IYS D E L DACWA+SYLSDGP + I+AV++ + RLVELLSH+S LVQTPALRAVGN
Sbjct 266 IYSMDTETLVDACWAISYLSDGPQEAIQAVIDVRIPKRLVELLSHESTLVQTPALRAVGN 325
Query 245 IVTGDDTQTQVVLTCGALPKLLTLLSSTKKNVQKEACWTVSNITAGNKDQIQEVLDSGAI 304
IVTG+D QTQVV+ G LP L LLSS K+N++KEACWT+SNITAGN +QIQ V+D+ I
Sbjct 326 IVTGNDLQTQVVINAGVLPALRLLLSSPKENIKKEACWTISNITAGNTEQIQAVIDANLI 385
Query 305 PALLHLLETADFDTKKEAAWAVSNAVSGGTDMQ--VALLVEKGCIPPLCSLLGCMDNKII 362
P L+ LLE A++ TKKEA WA+SNA SGG + LV +GCI PLC LL DN+II
Sbjct 386 PPLVKLLEVAEYKTKKEACWAISNASSGGLQRPDIIRYLVSQGCIKPLCDLLEIADNRII 445
Query 363 MVALEALNAILRTGKKIKETEGLSVNPYATLIEQADGATALEKLQDSTNENVFK 416
V L+AL IL+ G+ KE GL++N A IE+A G + Q + N+ +++
Sbjct 446 EVTLDALENILKMGEADKEARGLNINENADFIEKAGGMEKIFNCQQNENDKIYE 499
> At3g05720
Length=528
Score = 424 bits (1091), Expect = 1e-118, Method: Compositional matrix adjust.
Identities = 226/419 (53%), Positives = 291/419 (69%), Gaps = 11/419 (2%)
Query 8 QQLVQALASGVPDAELEGTQLFRRALSLEN-NPPIQEVIDAGAVPHFVRFLSMHGRYQLQ 66
Q+L+ + S D +E T R L E N ++EVI AG VP FV FL+ QLQ
Sbjct 62 QKLISCIWSDERDLLIEATTQIRTLLCGEMFNVRVEEVIQAGLVPRFVEFLTWDDSPQLQ 121
Query 67 FEAAWALTNIASGTQEQTRVVVECGAIPLFVELLRCPKEEVREQAVWALGNIAGDSPSCR 126
FEAAWALTNIASGT E T VV++ GA+ + V LL P + VREQ VWALGNI+GDSP CR
Sbjct 122 FEAAWALTNIASGTSENTEVVIDHGAVAILVRLLNSPYDVVREQVVWALGNISGDSPRCR 181
Query 127 NLVLDGGALPPLLEQLNTPGSKVAMLRNATWTLSNLCRGKPAPVFEKVSPALPALAVLIY 186
++VL ALP LL QLN G+K++ML NA WTLSNLCRGKP P F++VS ALPALA LI
Sbjct 182 DIVLGHAALPSLLLQLNH-GAKLSMLVNAAWTLSNLCRGKPQPPFDQVSAALPALAQLIR 240
Query 187 SDDAEVLTDACWALSYLSDGPNDRIKAVLEAGVCARLVELLSHQSNLVQTPALRAVGNIV 246
DD E+L CWAL YLSDG N++I+AV+EA VCARL+ L H+S V TPALR +GNIV
Sbjct 241 LDDKELLAYTCWALVYLSDGSNEKIQAVIEANVCARLIGLSIHRSPSVITPALRTIGNIV 300
Query 247 TGDDTQTQVVLTCGALPKLLTLL-SSTKKNVQKEACWTVSNITAGNKDQIQEVLDSGAIP 305
TG+D+QTQ ++ ALP L+ LL S K ++KEACWTVSNITAG + QIQ V D+ P
Sbjct 301 TGNDSQTQHIIDLQALPCLVNLLRGSYNKTIRKEACWTVSNITAGCQSQIQAVFDADICP 360
Query 306 ALLHLLETADFDTKKEAAWAVSNAVSGGTDMQVALLVEKGCIPPLCSLLGCMDNKIIMVA 365
AL++LL+ ++ D KKEAAWA+ NA++GG+ Q+ LV++ CI PLC LL C D +++MV
Sbjct 361 ALVNLLQNSEGDVKKEAAWAICNAIAGGSYKQIMFLVKQECIKPLCDLLTCSDTQLVMVC 420
Query 366 LEALNAILRTGKKI--KETEGL------SVNPYATLIEQADGATALEKLQDSTNENVFK 416
LEAL IL+ G+ + EG+ +VNP+A LIE+A+G +E LQ N ++++
Sbjct 421 LEALKKILKVGEVFSSRHAEGIYQCPQTNVNPHAQLIEEAEGLEKIEGLQSHENNDIYE 479
> At5g49310
Length=519
Score = 418 bits (1075), Expect = 1e-116, Method: Compositional matrix adjust.
Identities = 209/413 (50%), Positives = 279/413 (67%), Gaps = 4/413 (0%)
Query 5 VDLQQLVQALASGVPDAELEGTQLFRRALSLENNPPIQEVIDAGAVPHFVRFLSMHGRYQ 64
+++ ++ + S P +LE T FR LS + +PP VI +G VP FV FL +
Sbjct 70 LEIANMITGVFSDDPSLQLEYTTRFRVVLSFDRSPPTDNVIKSGVVPRFVEFLKKDDNPK 129
Query 65 LQFEAAWALTNIASGTQEQTRVVVECGAIPLFVELLRCPKEEVREQAVWALGNIAGDSPS 124
LQFEAAWALTNIASG E T+VV++ G +PLFV+LL P ++VREQA+W LGN+AGDS
Sbjct 130 LQFEAAWALTNIASGASEHTKVVIDHGVVPLFVQLLASPDDDVREQAIWGLGNVAGDSIQ 189
Query 125 CRNLVLDGGALPPLLEQLNTPGSKVAMLRNATWTLSNLCRGKPAPVFEKVSPALPALAVL 184
CR+ VL+ GA PLL QLN + +++LRNATWTLSN RGKP+P F+ V LP L L
Sbjct 190 CRDFVLNSGAFIPLLHQLNNHAT-LSILRNATWTLSNFFRGKPSPPFDLVKHVLPVLKRL 248
Query 185 IYSDDAEVLTDACWALSYLSDGPNDRIKAVLEAGVCARLVELLSHQSNLVQTPALRAVGN 244
+YSDD +VL DACWALS LSD N+ I++V+EAGV RLVELL H S +V PALR +GN
Sbjct 249 VYSDDEQVLIDACWALSNLSDASNENIQSVIEAGVVPRLVELLQHASPVVLVPALRCIGN 308
Query 245 IVTGDDTQTQVVLTCGALPKLLTLLSSTK-KNVQKEACWTVSNITAGNKDQIQEVLDSGA 303
IV+G+ QT V+ CG LP L LL+ + +++EACWT+SNITAG ++QIQ V+D+
Sbjct 309 IVSGNSQQTHCVINCGVLPVLADLLTQNHMRGIRREACWTISNITAGLEEQIQSVIDANL 368
Query 304 IPALLHLLETADFDTKKEAAWAVSNAVSGGTDMQVALLVEKGCIPPLCSLLGCMDNKIIM 363
IP+L++L + A+FD KKEA WA+SNA GG+ Q+ LVE+ CI LC +L C D +II+
Sbjct 369 IPSLVNLAQHAEFDIKKEAIWAISNASVGGSPNQIKYLVEQNCIKALCDILVCPDLRIIL 428
Query 364 VALEALNAILRTGKKIKETEGLSVNPYATLIEQADGATALEKLQDSTNENVFK 416
V+L L IL G+ K VN Y+ +IE A+G +E LQ N +++
Sbjct 429 VSLGGLEMILIAGEVDKNLR--DVNCYSQMIEDAEGLEKIENLQHHGNNEIYE 479
> Hs4504897
Length=529
Score = 397 bits (1020), Expect = 2e-110, Method: Compositional matrix adjust.
Identities = 212/414 (51%), Positives = 275/414 (66%), Gaps = 9/414 (2%)
Query 7 LQQLVQALASGVPDAELEGTQLFRRALSLENNPPIQEVIDAGAVPHFVRFLSMHGRYQLQ 66
+ +V+ + S + +L+ TQ R+ LS E PPI +I AG +P FV FL +Q
Sbjct 78 VDDIVKGINSSNVENQLQATQAARKLLSREKQPPIDNIIRAGLIPKFVSFLGRTDCSPIQ 137
Query 67 FEAAWALTNIASGTQEQTRVVVECGAIPLFVELLRCPKEEVREQAVWALGNIAGDSPSCR 126
FE+AWALTNIASGT EQT+ VV+ GAIP F+ LL P + EQAVWALGNIAGD R
Sbjct 138 FESAWALTNIASGTSEQTKAVVDGGAIPAFISLLASPHAHISEQAVWALGNIAGDGSVFR 197
Query 127 NLVLDGGALPPLLEQLNTPG-SKVA--MLRNATWTLSNLCRGK-PAPVFEKVSPALPALA 182
+LV+ GA+ PLL L P S +A LRN TWTLSNLCR K PAP + V LP L
Sbjct 198 DLVIKYGAVDPLLALLAVPDMSSLACGYLRNLTWTLSNLCRNKNPAPPIDAVEQILPTLV 257
Query 183 VLIYSDDAEVLTDACWALSYLSDGPNDRIKAVLEAGVCARLVELLSHQSNLVQTPALRAV 242
L++ DD EVL D CWA+SYL+DGPN+RI V++ GV +LV+LL + TPALRA+
Sbjct 258 RLLHHDDPEVLADTCWAISYLTDGPNERIGMVVKTGVVPQLVKLLGASELPIVTPALRAI 317
Query 243 GNIVTGDDTQTQVVLTCGALPKLLTLLSSTKKNVQKEACWTVSNITAGNKDQIQEVLDSG 302
GNIVTG D QTQVV+ GAL +LL++ K N+QKEA WT+SNITAG +DQIQ+V++ G
Sbjct 318 GNIVTGTDEQTQVVIDAGALAVFPSLLTNPKTNIQKEATWTMSNITAGRQDQIQQVVNHG 377
Query 303 AIPALLHLLETADFDTKKEAAWAVSNAVSGGTDMQVALLVEKGCIPPLCSLLGCMDNKII 362
+P L+ +L ADF T+KEA WAV+N SGGT Q+ LV G I PL +LL D KII
Sbjct 378 LVPFLVSVLSKADFKTQKEAVWAVTNYTSGGTVEQIVYLVHCGIIEPLMNLLTAKDTKII 437
Query 363 MVALEALNAILRTGKKIKETEGLSVNPYATLIEQADGATALEKLQDSTNENVFK 416
+V L+A++ I + +K+ ETE LS+ +IE+ G +E LQ+ NE+V+K
Sbjct 438 LVILDAISNIFQAAEKLGETEKLSI-----MIEECGGLDKIEALQNHENESVYK 486
> Hs4504901
Length=521
Score = 380 bits (975), Expect = 4e-105, Method: Compositional matrix adjust.
Identities = 196/416 (47%), Positives = 263/416 (63%), Gaps = 9/416 (2%)
Query 2 LEGVDLQQLVQALASGVPDAELEGTQLFRRALSLENNPPIQEVIDAGAVPHFVRFLSMHG 61
++ L+ +VQ +S +L Q R+ LS + NPPI ++I +G +P V L
Sbjct 68 VQNTSLEAIVQNASSDNQGIQLSAVQAARKLLSSDRNPPIDDLIKSGILPILVHCLERDD 127
Query 62 RYQLQFEAAWALTNIASGTQEQTRVVVECGAIPLFVELLRCPKEEVREQAVWALGNIAGD 121
LQFEAAWALTNIASGT EQT+ VV+ A+PLF+ LL P + V EQAVWALGNI GD
Sbjct 128 NPSLQFEAAWALTNIASGTSEQTQAVVQSNAVPLFLRLLHSPHQNVCEQAVWALGNIIGD 187
Query 122 SPSCRNLVLDGGALPPLLEQLNTPGSKVAMLRNATWTLSNLCRGK-PAPVFEKVSPALPA 180
P CR+ V+ G + PLL + +P + LRN TW + NLCR K P P E + LPA
Sbjct 188 GPQCRDYVISLGVVKPLLSFI-SPSIPITFLRNVTWVMVNLCRHKDPPPPMETIQEILPA 246
Query 181 LAVLIYSDDAEVLTDACWALSYLSDGPNDRIKAVLEAGVCARLVELLSHQSNLVQTPALR 240
L VLI+ D +L D WALSYL+D N++I+ V+++G+ LV LLSHQ VQT ALR
Sbjct 247 LCVLIHHTDVNILVDTVWALSYLTDAGNEQIQMVIDSGIVPHLVPLLSHQEVKVQTAALR 306
Query 241 AVGNIVTGDDTQTQVVLTCGALPKLLTLLSSTKKNVQKEACWTVSNITAGNKDQIQEVLD 300
AVGNIVTG D QTQVVL C AL LL+ K+ + KEA W +SNITAGN+ Q+Q V+D
Sbjct 307 AVGNIVTGTDEQTQVVLNCDALSHFPALLTHPKEKINKEAVWFLSNITAGNQQQVQAVID 366
Query 301 SGAIPALLHLLETADFDTKKEAAWAVSNAVSGGTDMQVALLVEKGCIPPLCSLLGCMDNK 360
+ +P ++HLL+ DF T+KEAAWA+SN G QVA L+++ IPP C+LL D +
Sbjct 367 ANLVPMIIHLLDKGDFGTQKEAAWAISNLTISGRKDQVAYLIQQNVIPPFCNLLTVKDAQ 426
Query 361 IIMVALEALNAILRTGKKIKETEGLSVNPYATLIEQADGATALEKLQDSTNENVFK 416
++ V L+ L+ IL+ + ET G LIE+ G +E+LQ+ NE+++K
Sbjct 427 VVQVVLDGLSNILKMAEDEAETIG-------NLIEECGGLEKIEQLQNHENEDIYK 475
> Hs4504899
Length=521
Score = 378 bits (971), Expect = 1e-104, Method: Compositional matrix adjust.
Identities = 200/415 (48%), Positives = 258/415 (62%), Gaps = 9/415 (2%)
Query 3 EGVDLQQLVQALASGVPDAELEGTQLFRRALSLENNPPIQEVIDAGAVPHFVRFLSMHGR 62
+ V L+ ++Q S P +L Q R+ LS + NPPI ++I +G +P V+ L
Sbjct 69 QNVTLEAILQNATSDNPVVQLSAVQAARKLLSSDQNPPIDDLIKSGILPILVKCLERDDN 128
Query 63 YQLQFEAAWALTNIASGTQEQTRVVVECGAIPLFVELLRCPKEEVREQAVWALGNIAGDS 122
LQFEAAWALTNIASGT QT+ VV+ A+PLF+ LLR P + V EQAVWALGNI GD
Sbjct 129 PSLQFEAAWALTNIASGTSAQTQAVVQSNAVPLFLRLLRSPHQNVCEQAVWALGNIIGDG 188
Query 123 PSCRNLVLDGGALPPLLEQLNTPGSKVAMLRNATWTLSNLCRGK-PAPVFEKVSPALPAL 181
P CR+ V+ G + PLL + +P + LRN TW + NLCR K P P E V LPAL
Sbjct 189 PQCRDYVISLGVVKPLLSFI-SPSIPITFLRNVTWVIVNLCRNKDPPPPMETVQEILPAL 247
Query 182 AVLIYSDDAEVLTDACWALSYLSDGPNDRIKAVLEAGVCARLVELLSHQSNLVQTPALRA 241
VLIY D +L D WALSYL+DG N++I+ V+++GV LV LLSHQ VQT ALRA
Sbjct 248 CVLIYHTDINILVDTVWALSYLTDGGNEQIQMVIDSGVVPFLVPLLSHQEVKVQTAALRA 307
Query 242 VGNIVTGDDTQTQVVLTCGALPKLLTLLSSTKKNVQKEACWTVSNITAGNKDQIQEVLDS 301
VGNIVTG D QTQVVL C L LLS K+ + KEA W +SNITAGN+ Q+Q V+D+
Sbjct 308 VGNIVTGTDEQTQVVLNCDVLSHFPNLLSHPKEKINKEAVWFLSNITAGNQQQVQAVIDA 367
Query 302 GAIPALLHLLETADFDTKKEAAWAVSNAVSGGTDMQVALLVEKGCIPPLCSLLGCMDNKI 361
G IP ++H L DF T+KEAAWA+SN G QV LV++ IPP C+LL D+++
Sbjct 368 GLIPMIIHQLAKGDFGTQKEAAWAISNLTISGRKDQVEYLVQQNVIPPFCNLLSVKDSQV 427
Query 362 IMVALEALNAILRTGKKIKETEGLSVNPYATLIEQADGATALEKLQDSTNENVFK 416
+ V L+ L IL G + A +IE+ G +E LQ NE+++K
Sbjct 428 VQVVLDGLKNILIMA-------GDEASTIAEIIEECGGLEKIEVLQQHENEDIYK 475
> CE20745
Length=514
Score = 359 bits (921), Expect = 7e-99, Method: Compositional matrix adjust.
Identities = 197/411 (47%), Positives = 258/411 (62%), Gaps = 10/411 (2%)
Query 7 LQQLVQALASGVPDAELEGTQLFRRALSLENNPPIQEVIDAGAVPHFVRFLSMHGRYQLQ 66
L+ V A S P +L Q R+ LS + NPPI ++I +G +P V+ LS LQ
Sbjct 67 LRLTVAAAQSSDPAEQLTAVQQARKMLSTDRNPPIDDLIGSGILPVLVQCLSSTDP-NLQ 125
Query 67 FEAAWALTNIASGTQEQTRVVVECGAIPLFVELLRCPKEEVREQAVWALGNIAGDSPSCR 126
FEAAWALTNIASGT EQT+ VV GA+PLF++LL C V EQ+VWALGNI GD P R
Sbjct 126 FEAAWALTNIASGTSEQTQAVVNAGAVPLFLQLLSCGNLNVCEQSVWALGNIIGDGPHFR 185
Query 127 NLVLDGGALPPLLEQLNTPGSKVAMLRNATWTLSNLCRGK-PAPVFEKVSPALPALAVLI 185
+ L+ G L PLL+ +N P + LRN TW + NLCR K PAP V LPAL++LI
Sbjct 186 DYCLELGILQPLLQFIN-PEIPIGFLRNVTWVIVNLCRCKDPAPSPAVVRTILPALSLLI 244
Query 186 YSDDAEVLTDACWALSYLSDGPNDRIKAVLEAGVCARLVELLSHQSNLVQTPALRAVGNI 245
+ D +L D WALSYL+DG N+ I+ V+EA V LV LL H VQT ALRAVGNI
Sbjct 245 HHQDTNILIDTVWALSYLTDGGNEHIQMVIEAQVVTHLVPLLGHVDVKVQTAALRAVGNI 304
Query 246 VTGDDTQTQVVLTCGALPKLLTLLSSTKKNVQKEACWTVSNITAGNKDQIQEVLDSGAIP 305
VTG D QTQ+VL G L + LL+ K+ + KEA W VSNITAGN+ Q+Q+V D+G +P
Sbjct 305 VTGTDEQTQLVLDSGVLRFMPGLLAHYKEKINKEAVWFVSNITAGNQQQVQDVFDAGIMP 364
Query 306 ALLHLLETADFDTKKEAAWAVSNAVSGGTDMQVALLVEKGCIPPLCSLLGCMDNKIIMVA 365
++HLL+ DF T+KEAAWA+SN G QV +V+ G + P C++L C D++II V
Sbjct 365 MIIHLLDRGDFPTQKEAAWAISNVTISGRPNQVEQMVKLGVLRPFCAMLSCTDSQIIQVV 424
Query 366 LEALNAILRTGKKIKETEGLSVNPYATLIEQADGATALEKLQDSTNENVFK 416
L+ +N IL+ + E + IE+ G +E LQ+ NE+++K
Sbjct 425 LDGINNILKMAGEAAEQ-------VTSEIEECGGLDKIENLQNHENEDIYK 468
> 7299211
Length=514
Score = 357 bits (915), Expect = 4e-98, Method: Compositional matrix adjust.
Identities = 187/399 (46%), Positives = 244/399 (61%), Gaps = 9/399 (2%)
Query 19 PDAELEGTQLFRRALSLENNPPIQEVIDAGAVPHFVRFLSMHGRYQLQFEAAWALTNIAS 78
P+ +L Q R+ LS + NPPI ++I + +P V L H LQFEAAWALTNIAS
Sbjct 80 PEQQLAAVQAARKLLSSDKNPPINDLIQSDILPILVECLKQHNHTMLQFEAAWALTNIAS 139
Query 79 GTQEQTRVVVECGAIPLFVELLRCPKEEVREQAVWALGNIAGDSPSCRNLVLDGGALPPL 138
GT QT VV GA+PLF++LL P V EQAVWALGNI GD P R+ V+ G + PL
Sbjct 140 GTSAQTNEVVAAGAVPLFLQLLNSPAPNVCEQAVWALGNIIGDGPLLRDFVIKHGVVQPL 199
Query 139 LEQLNTPGSKVAMLRNATWTLSNLCRGK-PAPVFEKVSPALPALAVLIYSDDAEVLTDAC 197
L + P + LRN TW + NLCR K PAP + LPAL VLI+ D +L D
Sbjct 200 LSFI-KPDIPITFLRNVTWVIVNLCRNKDPAPPTATIHEILPALNVLIHHTDTNILVDTV 258
Query 198 WALSYLSDGPNDRIKAVLEAGVCARLVELLSHQSNLVQTPALRAVGNIVTGDDTQTQVVL 257
WA+SYL+DG ND+I+ V+E+GV +L+ LL + VQT ALRAVGNIVTG D QTQVVL
Sbjct 259 WAISYLTDGGNDQIQMVIESGVVPKLIPLLGNSEVKVQTAALRAVGNIVTGSDEQTQVVL 318
Query 258 TCGALPKLLTLLSSTKKNVQKEACWTVSNITAGNKDQIQEVLDSGAIPALLHLLETADFD 317
AL LLS K+ ++KEA W +SNITAGN+ Q+Q V++ G +P ++ L +F
Sbjct 319 NYDALSYFPGLLSHPKEKIRKEAVWFLSNITAGNQSQVQAVINVGLLPKIIENLSKGEFQ 378
Query 318 TKKEAAWAVSNAVSGGTDMQVALLVEKGCIPPLCSLLGCMDNKIIMVALEALNAILRTGK 377
T+KEAAWA+SN G QV L+++G IPP C LL C D ++I V L+ LN +L+
Sbjct 379 TQKEAAWAISNLTISGNREQVFTLIKEGVIPPFCDLLSCQDTQVINVVLDGLNNMLKVAD 438
Query 378 KIKETEGLSVNPYATLIEQADGATALEKLQDSTNENVFK 416
V A IE+ +G +E+LQ N ++K
Sbjct 439 S-------HVEAVANCIEECEGLAKIERLQSHENVEIYK 470
> 7297600
Length=522
Score = 313 bits (801), Expect = 6e-85, Method: Compositional matrix adjust.
Identities = 187/412 (45%), Positives = 250/412 (60%), Gaps = 8/412 (1%)
Query 7 LQQLVQALASGVPDAELEGTQLFRRALSLENNPPIQEVIDAGAVPHFVRFLSMHGRYQLQ 66
+ ++V A+ S + + G Q R+ LS E NPPI +I G VP +RFL LQ
Sbjct 71 VDEIVAAMNSEDQERQFLGMQSARKMLSRERNPPIDLMIGHGIVPICIRFLQNTNNSMLQ 130
Query 67 FEAAWALTNIASGTQEQTRVVVECGAIPLFVELLRCPKEEVREQAVWALGNIAGDSPSCR 126
FEAAWALTNIASGT +QTR V+E A+P FV LL+ + EQAVWALGNIAGD + R
Sbjct 131 FEAAWALTNIASGTSDQTRCVIEHNAVPHFVALLQSKSMNLAEQAVWALGNIAGDGAAAR 190
Query 127 NLVLDGGALPPLLEQLNTPGSKVAMLRNATWTLSNLCRGK-PAPVFEKVSPALPALAVLI 185
++V+ + +L +N + ++ LRN W +SNLCR K P+P F++V LP L+ L+
Sbjct 191 DIVIHHNVIDGILPLINNE-TPLSFLRNIVWLMSNLCRNKNPSPPFDQVKRLLPVLSQLL 249
Query 186 YSDDAEVLTDACWALSYLSDGPNDRIKAVLEAGVCARLVELLSHQSNLVQTPALRAVGNI 245
S D +VL DACWALSY++D N +I+AV+++ RLV+LL + PALR+VGNI
Sbjct 250 LSQDIQVLADACWALSYVTDDDNTKIQAVVDSDAVPRLVKLLQMDEPSIIVPALRSVGNI 309
Query 246 VTGDDTQTQVVLTCGALPKLLTLLSSTKKNVQKEACWTVSNITAGNKDQIQEVLDSGAIP 305
VTG D QT VV+ G LP+L LL K N+ KEA WTVSNITAGN+ QIQ V+ +G
Sbjct 310 VTGTDQQTDVVIASGGLPRLGLLLQHNKSNIVKEAAWTVSNITAGNQKQIQAVIQAGIFQ 369
Query 306 ALLHLLETADFDTKKEAAWAVSNAVSGGTDMQVALLVEKGCI-PPLCSLLGCMDNKIIMV 364
L +LE DF +KEAAWAV+N + GT Q+ L+EK I P LL D + I V
Sbjct 370 QLRTVLEKGDFKAQKEAAWAVTNTTTSGTPEQIVDLIEKYKILKPFIDLLDTKDPRTIKV 429
Query 365 ALEALNAILRTGKKIKETEGLSVNPYATLIEQADGATALEKLQDSTNENVFK 416
L+ + +K+ TE L + ++E+ G LE LQ NE V+K
Sbjct 430 VQTGLSNLFALAEKLGGTENLCL-----MVEEMGGLDKLETLQQHENEEVYK 476
> At5g52000
Length=441
Score = 303 bits (775), Expect = 5e-82, Method: Compositional matrix adjust.
Identities = 170/409 (41%), Positives = 241/409 (58%), Gaps = 14/409 (3%)
Query 10 LVQALASGVPDAELEGTQLFRRALSLENNPPIQEVIDAGAVPHFVRFLSMHGRYQLQFEA 69
++ L S P +LE RR S + I VI +G VP V+ L +LQ+E
Sbjct 14 IIDGLWSDDPPLQLESVTKIRRITSQRD---ISCVIRSGVVPRLVQLLKNQVFPKLQYEV 70
Query 70 AWALTNIASGTQEQTRVVVECGAIPLFVELLRCPKEEVREQAVWALGNIAGDSPSCRNLV 129
AWALTNIA + ++L+ PK+ VREQA+W L N+AG S R+ V
Sbjct 71 AWALTNIAVDNPGVVVNNNAVPVL---IQLIASPKDYVREQAIWTLSNVAGHSIHYRDFV 127
Query 130 LDGGALPPLLEQLNTPGSKVAMLRNATWTLSNLCRGKPAPVFEKVSPALPALAVLIYSDD 189
L+ G L PLL L K LR ATW L NLCRGKP P F++V PALPAL +L++S D
Sbjct 128 LNSGVLMPLLRLL----YKDTTLRIATWALRNLCRGKPHPAFDQVKPALPALEILLHSHD 183
Query 190 AEVLTDACWALSYLSDGPNDRIKAVLEAGVCARLVELLSHQSNLVQTPALRAVGNIVTGD 249
+VL +AC AL +LS+G D I++V+EAG +LV++L S +V PAL +G + G+
Sbjct 184 EDVLKNACMALCHLSEGSEDGIQSVIEAGFVPKLVQILQLPSPVVLVPALLTIGAMTAGN 243
Query 250 DTQTQVVLTCGALPKLLTLLSSTKKN-VQKEACWTVSNITAGNKDQIQEVLDSGAIPALL 308
QTQ V+ GALP + +L+ +N ++K ACW +SNITAG K+QIQ V+D+ IP L+
Sbjct 244 HQQTQCVINSGALPIISNMLTRNHENKIKKCACWVISNITAGTKEQIQSVIDANLIPILV 303
Query 309 HLLETADFDTKKEAAWAVSNAVSGGTDMQVALLVEKGCIPPLCSLLGCMDNK-IIMVALE 367
+L + DF KKEA WA+SN G+ Q+ + E+ CI LC +L D + I+ L+
Sbjct 304 NLAQDTDFYMKKEAVWAISNMALNGSHDQIKYMAEQSCIKQLCDILVYSDERTTILKCLD 363
Query 368 ALNAILRTGKKIKETEGLSVNPYATLIEQADGATALEKLQDSTNENVFK 416
L +L+ G+ K +E VNPY LIE A+G + KLQ + N+++++
Sbjct 364 GLENMLKAGEAEKNSE--DVNPYCLLIEDAEGLEKISKLQMNKNDDIYE 410
> CE09675
Length=531
Score = 260 bits (664), Expect = 4e-69, Method: Compositional matrix adjust.
Identities = 153/412 (37%), Positives = 235/412 (57%), Gaps = 14/412 (3%)
Query 11 VQALASGVP--DAELEGTQLFRRALSLENNPPIQEVIDAGAVPHFVRFLSMHGRYQLQFE 68
++A+ S P D + + R++LS NPPI EVI G + V+ LS+ ++Q+E
Sbjct 93 IRAILSNNPSEDDMVRCFESLRKSLSKTKNPPIDEVIHCGLLQALVQALSVENE-RVQYE 151
Query 69 AAWALTNIASGTQEQTRVVVECGAIPLFVELLRCPKEEVREQAVWALGNIAGDSPSCRNL 128
AAWALTNI SG+ EQT VE G + L ++ EQA+WA+ NIAGDS R+
Sbjct 152 AAWALTNIVSGSTEQTIAAVEAGVTIPLIHLSVHQSAQISEQALWAVANIAGDSSQLRDY 211
Query 129 VLDGG---ALPPLLEQLNTPGSKVAMLRNATWTLSNLCRGK-PAPVFEKVSPALPALAVL 184
V+ AL L+E+++ G + +R W SN+CR K P E + L L
Sbjct 212 VIKCHGVEALMHLMEKVDQLGD--SHVRTIAWAFSNMCRHKNPHAPLEVLRVLSKGLVKL 269
Query 185 IYSDDAEVLTDACWALSYLSDGPNDRIKAVLEAGVCARLVELLSHQSNLVQTPALRAVGN 244
+ D +V DACWA+SYL+DGP+++I+ E+GV +V NLV PALR +GN
Sbjct 270 VQHTDRQVRQDACWAVSYLTDGPDEQIELARESGVLPHVVAFFKEAENLV-APALRTLGN 328
Query 245 IVTGDDTQTQVVLTCGALPKLLTLLSSTKKN-VQKEACWTVSNITAGNKDQIQEVLDSGA 303
+ TG+D+ TQ V+ G+L ++L L+ T+ + + KE CW VSNI AG + QIQ VLD+
Sbjct 329 VATGNDSLTQAVIDLGSLDEILPLMEKTRSSSIVKECCWLVSNIIAGTQKQIQAVLDANL 388
Query 304 IPALLHLLETADFDTKKEAAWAVSNAVSGGTDMQVALLVEKGCIPPLCSLLGCMDNKIIM 363
+P L+++L++ D + EA+WA+SN GGT+ QV ++E +P LC L + ++
Sbjct 389 LPVLINVLKSGDHKCQFEASWALSNLAQGGTNRQVVAMLEDNVVPALCQALLQTNTDMLN 448
Query 364 VALEALNAILRTGKKIKETEGLSVNPYATLIEQADGATALEKLQDSTNENVF 415
LE L ++ T ++ V+ +E+ G +LE+LQ+S +E ++
Sbjct 449 NTLETLYTLMLT---VQNGYPHKVDILHDQVEENGGLDSLERLQESQSEQIY 497
> ECU11g1760
Length=537
Score = 232 bits (591), Expect = 1e-60, Method: Compositional matrix adjust.
Identities = 156/455 (34%), Positives = 235/455 (51%), Gaps = 66/455 (14%)
Query 24 EGTQLFRRALSLENNPPIQEVIDAGAVPHFVR-----FLSMHGRY--------------- 63
+G FR LS+E++PPIQ VID+G VP F+ FL+ G
Sbjct 68 QGVYEFRSLLSVEDSPPIQPVIDSGCVPRFIELMDISFLTSSGMVMGGIPVSNIQRDMPE 127
Query 64 ---------QLQFEAAWALTNIASGTQEQTRVVVECGAIPLFVELLRCPKEEVREQAVWA 114
+++ EAAW +TNI+SGT +QT+VV++CGA+PL +++L + V +Q+VWA
Sbjct 128 FSVAMDIINKIRVEAAWVITNISSGTTQQTKVVIDCGAVPLLIQMLLDTNDAVLDQSVWA 187
Query 115 LGNIAGDSPSCRNLVLDGGALPP---LLEQLNTPGSKVAMLRNATWTLSNLCRGK-PAPV 170
LGNIAGDS R+++L+ GAL LL +L+ + ++RN TW LSNL RG+ P P
Sbjct 188 LGNIAGDSEGMRDIILETGALDTVLGLLLKLHESAIHIKIVRNLTWLLSNLNRGRNPPPS 247
Query 171 FEKVSPALPALAVLIYSDDAEVLTDACWALSYLSDGPNDRIKAVLEAGVCARLVELLSHQ 230
+ + +L L LI D EV++DA WALSY+ D + VL +G R LL
Sbjct 248 EQNMRKSLEILFKLINIRDPEVVSDAYWALSYIVDVSMECSDIVLNSGTMGRTHSLLEAL 307
Query 231 SNLVQ----------------TPALRAVGNIVTGDDTQTQVVLTCGAL----PKLLTLLS 270
+N + +P +R +GNIVTG D QT ++ G L P L +
Sbjct 308 TNRLVGMSYSAEDAKICLCAISPIIRMIGNIVTGSDDQTSALIRMGFLKFFRPIFYRLEN 367
Query 271 STKKNVQKEACWTVSNITAGNKDQIQEVLDSGAIPALLHLLETADFDTKKEAAWAV---- 326
+ ++KE CWT+SNITAG + + V+D P L+ + + + +KEA WAV
Sbjct 368 KKQPRLRKEICWTLSNITAGTVEHAKAVMDMDLAPLLIDSMSSYELFIRKEACWAVLNLM 427
Query 327 --SNAVSGGTDMQ---VALLVEKGCIPPLCSLLGCMDN--KIIMVALEALNAILRTGKKI 379
N + +D++ + +L++ G I L S L + N ++ L+ L GK+
Sbjct 428 YHCNNLPSSSDIKFEWLKILMDNGFIDALESYLDVVPNVPEMQCQILDCFRYALNGGKQW 487
Query 380 KETEGLSVNPYATLIEQADGATALEKLQDSTNENV 414
+ G NP + +AD A+E LQDS + V
Sbjct 488 ELRYG--KNPIYEKMVEADIVDAIEGLQDSVDRTV 520
> Hs17443779
Length=372
Score = 156 bits (394), Expect = 8e-38, Method: Compositional matrix adjust.
Identities = 90/181 (49%), Positives = 109/181 (60%), Gaps = 4/181 (2%)
Query 103 PKEEVREQAVWALGNIAGDSPSCRNLVLDGGALPPLLEQLNTPGSK---VAMLRNATWTL 159
P + EQAVWALGNIAGD R+LV+ GA+ PLL L P L + TWTL
Sbjct 95 PHAHISEQAVWALGNIAGDGLVFRDLVIKYGAVDPLLALLAVPDMSSLVCGYLCHLTWTL 154
Query 160 SNLCRGK-PAPVFEKVSPALPALAVLIYSDDAEVLTDACWALSYLSDGPNDRIKAVLEAG 218
S LC K PAP + V L L L++ DD EVL D+CWA+SYL+DGPN R L+ G
Sbjct 155 SILCCYKNPAPQLDAVEQILSTLVWLLHHDDPEVLADSCWAISYLTDGPNKRTDMFLKIG 214
Query 219 VCARLVELLSHQSNLVQTPALRAVGNIVTGDDTQTQVVLTCGALPKLLTLLSSTKKNVQK 278
V +LV+LL TPALR +GNIVTG D +TQVV+ GAL LL ++K N QK
Sbjct 215 VLPQLVKLLGASELPTVTPALRPIGNIVTGTDEETQVVIDAGALAVFPNLLINSKTNFQK 274
Query 279 E 279
E
Sbjct 275 E 275
Score = 36.2 bits (82), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 20/80 (25%), Positives = 41/80 (51%), Gaps = 0/80 (0%)
Query 56 FLSMHGRYQLQFEAAWALTNIASGTQEQTRVVVECGAIPLFVELLRCPKEEVREQAVWAL 115
+L H ++ ++ WA++ + G ++T + ++ G +P V+LL + A+ +
Sbjct 179 WLLHHDDPEVLADSCWAISYLTDGPNKRTDMFLKIGVLPQLVKLLGASELPTVTPALRPI 238
Query 116 GNIAGDSPSCRNLVLDGGAL 135
GNI + +V+D GAL
Sbjct 239 GNIVTGTDEETQVVIDAGAL 258
Score = 32.0 bits (71), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 21/65 (32%), Positives = 35/65 (53%), Gaps = 5/65 (7%)
Query 48 GAVPHFVRFLSMHGRYQLQF--EAAWALTNIASGTQEQTRVVVECGAIPLFVELLRCPKE 105
G +P V+ L G +L A + NI +GT E+T+VV++ GA+ +F LL K
Sbjct 214 GVLPQLVKLL---GASELPTVTPALRPIGNIVTGTDEETQVVIDAGALAVFPNLLINSKT 270
Query 106 EVREQ 110
+++
Sbjct 271 NFQKE 275
> 7295403
Length=442
Score = 155 bits (391), Expect = 2e-37, Method: Compositional matrix adjust.
Identities = 104/378 (27%), Positives = 193/378 (51%), Gaps = 12/378 (3%)
Query 39 PPIQEVIDAGAVPHFVRFLSMHGRYQLQFEAAWALTNIASGTQEQTRVVVECGAIPLFVE 98
P + +++ + + + V L + +++ +AA AL +IASG+ E + ++ + GA+P +
Sbjct 53 PDVNKLLKSNTIGNLVESLGHGNKNKIRADAADALAHIASGSSEHSNLIAKAGAVPRLIR 112
Query 99 LLRCPKEEVREQAVWALGNIAGDSPSCRNLVLDGGALPPLLEQLNTPGSKVAMLRNATWT 158
LL+ P EV E+ + +LGN+ +P+ R+ ++ G + L+ + + ML + TW
Sbjct 113 LLQSPDPEVCEKGILSLGNLLHFAPNLRDYIIRHGLMQKLMSIIQDKSTCTLMLSHVTWV 172
Query 159 LSNLC-RGKPAPVFEKVSPALPALAVLIYSDDAEVLTDACWALSYLSDGPNDRIKAVLEA 217
L LC +P+P + + + AL +++Y+ +A VL DA A+ L+ G N+ I+ +L+
Sbjct 173 LRKLCISSQPSPP-DNAAEIIQALNIVLYNPEANVLEDALMAVRNLAHG-NETIQMLLDF 230
Query 218 GVCARLVELLSHQSNLVQTPALRAVGNIVTGDDTQTQVVLTCGALPKLLTLLSSTKKNVQ 277
V R++ LL H + VQ AL+A+ NI TG + Q Q +L LP L L+S++ +++
Sbjct 231 EVVPRIIYLLEHPNVTVQNAALQALINIATGSEEQIQELLNNNLLPHLSALMSNSDPDIR 290
Query 278 KEACWTVSNITAGNKDQIQEVLDSGAIPALLHLLETADFDTKKEAAWAVSNAVSGGTDMQ 337
+ + NI GN Q ++++G + +L L+ K AA ++
Sbjct 291 CQVLKLLLNIADGNIFQRHAIMNAGLLHKILECLKADAISLKSAAALTITTLAIDKDKNL 350
Query 338 VALLVEKGCIPPLCSLLGCMDNKIIMVALEALNAILRTGKKIK-ETEGLSVNPYATLIEQ 396
+ L+ +G IP C+LL C + I+ L+ L+ +L E G +IE
Sbjct 351 LCYLMRQGVIPEFCNLLFCQERDILSNVLDILSTMLDVDPSFSAEVSG--------IIEW 402
Query 397 ADGATALEKLQDSTNENV 414
+ + LQ S +E +
Sbjct 403 SGALNNIRMLQSSEHEEI 420
> At5g03070
Length=483
Score = 150 bits (379), Expect = 5e-36, Method: Compositional matrix adjust.
Identities = 118/421 (28%), Positives = 191/421 (45%), Gaps = 53/421 (12%)
Query 6 DLQQLVQALASGVPDAELEGTQLFRRALSLENNPPIQEVIDAGAVPHFVRFLSMHGRYQL 65
+L+ VQ G + + RR LS PP++ + AGA+P V+ LS +
Sbjct 84 ELKSAVQYQGKGAMQKRVTALRELRRLLSKSEFPPVEAALRAGAIPLLVQCLSFGSPDEQ 143
Query 66 QFEAAWALTNIASGTQEQTRVVVECGAIPLFVELLRCPKEEVREQAVWALGNIAGDSPSC 125
E+AW LTNIA+G E+T+ ++ + + L V EQ WA+GN+AG+
Sbjct 144 LLESAWCLTNIAAGKPEETKALLPALPLLI-AHLGEKSSAPVAEQCAWAIGNVAGEGEDL 202
Query 126 RNLVLDGGALPPLLEQLNTPGSKVAMLRNATWTLSNLCRG---KPAPVFEKVSPALPALA 182
RN++L GALPPL + K + +R A W LSNL +G K A K+ L A+
Sbjct 203 RNVLLSQGALPPLARMIFP--DKGSTVRTAAWALSNLIKGPESKAAAQLVKIDGILDAIL 260
Query 183 VLIYSDDAEVLTDACWALSYLSDGPNDRIKAVLEAGVCARLVELLSHQSNLVQTPALRAV 242
+ D E T+ W + YLS + +L+ G+ L++ L+ S+L
Sbjct 261 RHLKKTDEETATEIAWIIVYLSALSDIATSMLLKGGILQLLIDRLATSSSL--------- 311
Query 243 GNIVTGDDTQTQVVLTCGALPKLLTLLSSTKKNVQKEACWTVSNITAGNKDQIQEVLDSG 302
Q+++ C L S + ++KEA W +SNI AG+ + + + +
Sbjct 312 -----------QLLIPC---------LRSEHRVLKKEAAWVLSNIAAGSIEHKRMIHSTE 351
Query 303 AIPALLHLLETADFDTKKEAAWAVSN--AVSGGTDMQVALLVEK-------GCIPPLCSL 353
+P LL +L T+ FD +KE A+ + N S D + ++ E GC+ L
Sbjct 352 VMPLLLRILSTSPFDIRKEVAYVLGNLCVESAEGDRKPRIIQEHLVSIVSGGCLRGFIEL 411
Query 354 LGCMDNKIIMVALEALNAILRTGKKIKETEGLSVNPYATLIEQADGATALEKLQDSTNEN 413
+ D + + L+ + +LR G+ L+E DG A+E+ Q NE
Sbjct 412 VRSPDIEAARLGLQFIELVLR---------GMPNGEGPKLVEGEDGIDAMERFQFHENEE 462
Query 414 V 414
+
Sbjct 463 L 463
> Hs22051967_1
Length=453
Score = 124 bits (310), Expect = 6e-28, Method: Compositional matrix adjust.
Identities = 97/306 (31%), Positives = 137/306 (44%), Gaps = 55/306 (17%)
Query 111 AVWALGNIAGDSPSCRNLVLDGGALPPLLEQLNTPGSKVAMLRNATWTLSNLCRGK-PAP 169
A+WALGNI D R+ V+ A+P L L +P + +RN TWT SNLCR K P P
Sbjct 59 AMWALGNIPSDGLEFRDNVISSNAIPHL-RALISPTLPITFMRNITWTWSNLCRKKTPYP 117
Query 170 VFEKVSPALPALAVLIYSDDAEVLTDACWALSYLSDGPNDRIKAVLEAGVCARLVELLSH 229
V L AL+ L+ +EVL++ACW+LSYL+ G C L+ +
Sbjct 118 CETAVKQMLRALS-LLQRHHSEVLSEACWSLSYLTYG-------------CKELIGQVDD 163
Query 230 QSNLVQTPAL-RAVGNIVTGDDTQTQVVLTCGALPKLLTLLSSTKKNVQKEACWTVSNIT 288
N + P L R +GNI+TG + Q Q+ + G L L L K ++Q EA W +SN+
Sbjct 164 TGNSMSGPLLSRTMGNIITGTNEQKQMAIDAGMLNVLPQFLQHNKSSIQ-EAAWALSNVA 222
Query 289 AGNKDQIQEVLDSGAIP---ALLHLLETADFDTKKEAAWAVSNAVSGGTDMQVALLVEKG 345
AG Q++L +P ALL +E + W V
Sbjct 223 AGPSHHTQQLLAYNVLPPVVALLKKMENLKSRKRLSGRWLTLQYV--------------- 267
Query 346 CIPPLCSLLGCMDNKIIMVALEALNAILRTGKKIKETEGLSVNPYATLIEQADGATALEK 405
KI+++ L+ ++ IL+ +K E E L LIEQ G +E
Sbjct 268 --------------KIVLIILDVISCILQAAEKRSEKEILCF-----LIEQLGGIDRIEA 308
Query 406 LQDSTN 411
LQ N
Sbjct 309 LQLHEN 314
Score = 39.3 bits (90), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 23/60 (38%), Positives = 35/60 (58%), Gaps = 2/60 (3%)
Query 42 QEVIDAGAVPHFVRFLSMHGRYQLQFEAAWALTNIASGTQEQTRVVVECGAIPLFVELLR 101
Q IDAG + +FL H + +Q EAAWAL+N+A+G T+ ++ +P V LL+
Sbjct 189 QMAIDAGMLNVLPQFLQ-HNKSSIQ-EAAWALSNVAAGPSHHTQQLLAYNVLPPVVALLK 246
> Hs20538368
Length=317
Score = 114 bits (285), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 75/235 (31%), Positives = 126/235 (53%), Gaps = 12/235 (5%)
Query 182 AVLIYSDDAEVLTDACWALSYLSDGPNDRIKAVLEAGVCARLVELLSHQSNLVQTPALRA 241
+V+ +D + + ++ WAL+ ++ G ++ KAV++ G + LL+ + A+ A
Sbjct 57 SVVKRTDCSPIQFESAWALTNIASGALEQTKAVVDGGAIPAFIFLLASPHAHISEQAVWA 116
Query 242 VGNIVTGDDTQTQVVLTCGALPKLLTLLSSTKKNVQKEACWTVSNITAGNKDQIQEVLDS 301
VGNI +V+ CGA L ++ AC + G++DQI++V++
Sbjct 117 VGNIAGDGSIFRDLVINCGA--VDALLALLAVPDMSSLAC----DYLPGHQDQIEQVVNH 170
Query 302 GAIPALLHLLETADFDTKKEAAWAVSNAVSGGTDMQVALLVEKGCIPPLCSLLGCMDNKI 361
G +P +++L ADF TKKE+ WAV+N S GT Q+ V G I PL +LL D K
Sbjct 171 GLVPFFVNVLSKADFKTKKESVWAVTNYSSSGTVEQIVYPVHCGIIKPLMNLLTARDTKF 230
Query 362 IMVALEALNAILRTGKKIKETEGLSVNPYATLIEQADGATALEKLQDSTNENVFK 416
I+V L+A++ I + +K+ ETE S++ ++ G +E LQ+ NE+V K
Sbjct 231 ILVILDAISNIFQAPEKLGETEKPSMS------KECGGLEKIEALQNHENESVCK 279
Score = 99.8 bits (247), Expect = 9e-21, Method: Compositional matrix adjust.
Identities = 88/285 (30%), Positives = 135/285 (47%), Gaps = 39/285 (13%)
Query 20 DAELEGTQLFRRALSLENNPPIQEVIDAGAVPH-FVRFLSMHGRYQ-------------- 64
D+ G QL RA + ++E+ A P +VR LS +Y+
Sbjct 8 DSNWTGWQLAIRATA-----KLEELRQAEIYPQAYVRHLSNIQKYKGDLFRGLKSVVKRT 62
Query 65 ----LQFEAAWALTNIASGTQEQTRVVVECGAIPLFVELLRCPKEEVREQAVWALGNIAG 120
+QFE+AWALTNIASG EQT+ VV+ GAIP F+ LL P + EQAVWA+GNIAG
Sbjct 63 DCSPIQFESAWALTNIASGALEQTKAVVDGGAIPAFIFLLASPHAHISEQAVWAVGNIAG 122
Query 121 DSPSCRNLVLDGGALPPLLEQLNTPGSKVAMLRNATWTLSNLCRGKPAPVFEKVSPAL-P 179
D R+LV++ GA+ LL L P + + + G + + V+ L P
Sbjct 123 DGSIFRDLVINCGAVDALLALLAVP--------DMSSLACDYLPGHQDQIEQVVNHGLVP 174
Query 180 ALAVLIYSDDAEVLTDACWALS-YLSDGPNDRIKAVLEAGVCARLVELLSHQSNLVQTPA 238
++ D + ++ WA++ Y S G ++I + G+ L+ LL+ +
Sbjct 175 FFVNVLSKADFKTKKESVWAVTNYSSSGTVEQIVYPVHCGIIKPLMNLLTARDTKFILVI 234
Query 239 LRAVGNIVT-----GDDTQTQVVLTCGALPKLLTLLSSTKKNVQK 278
L A+ NI G+ + + CG L K+ L + ++V K
Sbjct 235 LDAISNIFQAPEKLGETEKPSMSKECGGLEKIEALQNHENESVCK 279
> Hs17469107
Length=246
Score = 113 bits (282), Expect = 9e-25, Method: Compositional matrix adjust.
Identities = 74/198 (37%), Positives = 109/198 (55%), Gaps = 32/198 (16%)
Query 215 LEAGVCARLVELLSHQSNLVQTPALRAVGNIVTGDDTQTQVVLTCGALPKLLTLLSSTKK 274
+E GV +LV+LL + TPALRA+GNIVTG QTQVV+ AL +LL+ K
Sbjct 1 METGVVPQLVKLLGASELPIVTPALRAIGNIVTGTGEQTQVVIDAEALAVFPSLLTDPKT 60
Query 275 NVQKEACWTVSNITAGNKDQIQEVLDSGAIPALLHLLETADFDTKKEAAWAVSNAVSGGT 334
N KEA WT+SNITAG +DQIQ+V++ G + L ++L +K
Sbjct 61 NTWKEAMWTMSNITAGRQDQIQQVVNHGLVLFLANVLSQILRHKRK-------------- 106
Query 335 DMQVALLVEKGCIPPLCSLLGCMDNKIIMVALEALNAILRTGKKIKETEGLSVNPYATLI 394
L +++ LL C D K+I+V L+A+++I + +K+ ETE LS+ +I
Sbjct 107 -----LTIDE--------LLNCKDTKVILVILDAISSIFQAAEKLGETEKLSI-----MI 148
Query 395 EQADGATALEKLQDSTNE 412
E+ G +E LQ+ N+
Sbjct 149 EECGGLDKIEALQNHEND 166
Score = 48.5 bits (114), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 30/89 (33%), Positives = 47/89 (52%), Gaps = 2/89 (2%)
Query 45 IDAGAVPHFVRFLSMHGRYQLQFEAAWALTNIASGTQEQTRVVVECGAIPLFVELLRCPK 104
++ G VP V+ L + A A+ NI +GT EQT+VV++ A+ +F LL PK
Sbjct 1 METGVVPQLVKLLGA-SELPIVTPALRAIGNIVTGTGEQTQVVIDAEALAVFPSLLTDPK 59
Query 105 EEVREQAVWALGNI-AGDSPSCRNLVLDG 132
++A+W + NI AG + +V G
Sbjct 60 TNTWKEAMWTMSNITAGRQDQIQQVVNHG 88
Score = 37.4 bits (85), Expect = 0.062, Method: Compositional matrix adjust.
Identities = 26/91 (28%), Positives = 43/91 (47%), Gaps = 2/91 (2%)
Query 88 VECGAIPLFVELLRCPKEEVREQAVWALGNIAGDSPSCRNLVLDGGALPPLLEQLNTPGS 147
+E G +P V+LL + + A+ A+GNI + +V+D AL L P
Sbjct 1 METGVVPQLVKLLGASELPIVTPALRAIGNIVTGTGEQTQVVIDAEALAVFPSLLTDP-- 58
Query 148 KVAMLRNATWTLSNLCRGKPAPVFEKVSPAL 178
K + A WT+SN+ G+ + + V+ L
Sbjct 59 KTNTWKEAMWTMSNITAGRQDQIQQVVNHGL 89
Score = 32.0 bits (71), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 20/59 (33%), Positives = 30/59 (50%), Gaps = 1/59 (1%)
Query 42 QEVIDAGAVPHFVRFLSMHGRYQLQFEAAWALTNIASGTQEQTRVVVECGAIPLFVELL 100
Q VIDA A+ F L+ + EA W ++NI +G Q+Q + VV G + +L
Sbjct 40 QVVIDAEALAVFPSLLT-DPKTNTWKEAMWTMSNITAGRQDQIQQVVNHGLVLFLANVL 97
> Hs17464994
Length=438
Score = 112 bits (280), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 87/262 (33%), Positives = 128/262 (48%), Gaps = 55/262 (20%)
Query 158 TLSNLCRGK-PAPVFEKVSPALPALAVLIYSDDAEVLTDACWALSYLSDGPNDRIKAVLE 216
TL+NLC K PA V LP L L++ ++ EVL D CWA+SYL+DG N+ I V++
Sbjct 187 TLTNLCCNKNPAAPLGAVEKILPTLVWLLHHNNPEVLVDTCWAVSYLTDGLNEWIDMVVK 246
Query 217 AGVCARLVELLSHQSNLVQTPALRAVGNIVT-GDDTQTQVVLTCGALPKLLTLLSSTKKN 275
V +LV+LL + TPALRA+GNIVT G Q Q + +L+++LS
Sbjct 247 TEVVTQLVKLLGVSELPIVTPALRAIGNIVTAGLQDQIQQAMN----HRLVSVLSKADFK 302
Query 276 VQKEACWTVSNIT-AGNKDQIQEVLDSGAIPALLHLLETADFDTKKEAAWAVSNAVSGGT 334
QKEA V+N T +G +QI ++ G I L++LL
Sbjct 303 TQKEAVLAVTNSTSSGIVEQIVYLVHCGIIEPLMNLLTVK-------------------- 342
Query 335 DMQVALLVEKGCIPPLCSLLGCMDNKIIMVALEALNAILRTGKKIKETEGLSVNPYATLI 394
D KII+V L+A++ I + +K+ ETE LS+ +I
Sbjct 343 -----------------------DTKIILVTLDAISNIFQAVEKLGETEKLSI-----MI 374
Query 395 EQADGATALEKLQDSTNENVFK 416
E+ G +E LQ+ N+ ++K
Sbjct 375 EECGGLDKIEALQNHANQFIYK 396
> CE28029
Length=447
Score = 96.3 bits (238), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 80/310 (25%), Positives = 138/310 (44%), Gaps = 17/310 (5%)
Query 66 QFEAAWALTNIASGTQEQTRVVVECGAIPLFVELLRCPKEEVREQAVWALGNIAGDSPSC 125
+ A W +TN+ +QE T + V + + ++L+R + Q VWA+ NIA D SC
Sbjct 58 EVNAVWIITNMCCISQEVTHLFVNSNCLDILIKLVRSQNARLSSQTVWAIANIAADCSSC 117
Query 126 RNLVLDGGALPPLLEQLNTPGS-KVAMLRNATWTLSNLCRG----KPAPVFEKVSPALPA 180
+ L L L T A R W ++N+ G P+PV + AL
Sbjct 118 KLKCRSPKLLKILARNLQTSYKLDDADRRQLIWCVNNIFSGGRTTMPSPVARSLICALS- 176
Query 181 LAVLIYSDDAEVL---TDACWALSYLSDGPND--RIKAVLEAG-VCARLVELLSHQSNL- 233
A+L+ +D + + + W L+ L D D RI +L V ++E Q++
Sbjct 177 -AILLDTDAVDNMGCGSMILWTLANLVDNSLDTTRIDMLLSQNYVVEHILEKFIDQTDKS 235
Query 234 VQTPALRAVGNIVTGDDTQTQVVLTCGALPKLLTLLSSTKKNVQKEACWTVSNITAGNKD 293
LR +GNI G+D QT +++ ++ + S+ ++ EA W +SN+ AG
Sbjct 236 CNVSCLRLIGNIAVGNDVQTDLLVDNPKFRVVMNIAMSSPEH-HSEAAWIISNVVAGAPR 294
Query 294 QIQEVLD--SGAIPALLHLLETADFDTKKEAAWAVSNAVSGGTDMQVALLVEKGCIPPLC 351
+ ++D ++ + T D +KE W + N ++ + Q LLV G +
Sbjct 295 HVDYIMDDPDAFYEWVMAGIHTGDKRLRKECLWIIGNLLATADEHQTYLLVSLGIVQHFP 354
Query 352 SLLGCMDNKI 361
+LL D ++
Sbjct 355 ALLDFDDVRL 364
> At5g13060
Length=706
Score = 84.7 bits (208), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 85/305 (27%), Positives = 136/305 (44%), Gaps = 24/305 (7%)
Query 68 EAAWALTNIASGTQEQTRVVVECGAIPLFVELLRCP-----------KEEVREQAVWALG 116
EAA + ++A E ++VE GAIP V L P + ++ + ALG
Sbjct 72 EAAADIADLAK-IDENVEIIVENGAIPALVRYLESPLVVCGNVPKSCEHKLEKDCALALG 130
Query 117 NIAGDSPSCRNLVLDGGALPPLLEQLNTPGS------KVAMLRNATWTLSNLCRGKPA-P 169
IA P + L++D GA+ P ++ L G A++R A ++N+ P
Sbjct 131 LIAAIQPGYQQLIVDAGAIVPTVKLLKRRGECGECMFANAVIRRAADIITNIAHDNPRIK 190
Query 170 VFEKVSPALPALAVLIYSDDAEVLTDACWALSYLSDGPNDRIKAVLEAGVCARLVELLSH 229
+V + L L+ D +V A AL +S ND K+ L A LV +L
Sbjct 191 TNIRVEGGIAPLVELLNFPDVKVQRAAAGALRTVS-FRNDENKSQLNA--LPTLVLMLQS 247
Query 230 QSNLVQTPALRAVGNIVTGDDTQTQVVLTCGALPKLLTLLSSTKKNVQKEACWTVSNITA 289
Q + V A+ A+GN+V + V+ GAL ++ LLSST Q+EA + A
Sbjct 248 QDSTVHGEAIGAIGNLVHSSPDIKKEVIRAGALQPVIGLLSSTCLETQREAALLIGQFAA 307
Query 290 GNKDQIQEVLDSGAIPALLHLLETADFDTKKEAAWAVSNAVSGGTDMQVALLVEKGCIPP 349
+ D + GAI L+ +LE++D + +A+A+ + A + +G I
Sbjct 308 PDSDCKVHIAQRGAITPLIKMLESSDEQVVEMSAFALGRLAQDAHNQ--AGIAHRGGIIS 365
Query 350 LCSLL 354
L +LL
Sbjct 366 LLNLL 370
Score = 37.0 bits (84), Expect = 0.084, Method: Compositional matrix adjust.
Identities = 37/129 (28%), Positives = 62/129 (48%), Gaps = 5/129 (3%)
Query 35 LENNPPIQ-EVIDAGAVPHFVRFLSMHGRYQLQFEAAWALTNIASGTQEQTRVVVECGAI 93
+ ++P I+ EVI AGA+ + LS + Q EAA + A+ + + + GAI
Sbjct 264 VHSSPDIKKEVIRAGALQPVIGLLSSTC-LETQREAALLIGQFAAPDSDCKVHIAQRGAI 322
Query 94 PLFVELLRCPKEEVREQAVWALGNIAGDSPSCRNLVLDGGALPPLLEQLNTPGSKVAMLR 153
+++L E+V E + +ALG +A D+ + + GG + LL L+ V
Sbjct 323 TPLIKMLESSDEQVVEMSAFALGRLAQDAHNQAGIAHRGGII-SLLNLLDVKTGSVQ--H 379
Query 154 NATWTLSNL 162
NA + L L
Sbjct 380 NAAFALYGL 388
> At5g19330
Length=636
Score = 84.3 bits (207), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 72/277 (25%), Positives = 129/277 (46%), Gaps = 20/277 (7%)
Query 69 AAWALTNIASGTQEQTRVVVECGAIPLFVELLRCP------------KEEVREQAVWALG 116
A L +A ++ V+V+ GA+P + L+ P + EV + + +ALG
Sbjct 74 ATQVLAELAKNAEDLVNVIVDGGAVPALMTHLQAPPYNDGDLAEKPYEHEVEKGSAFALG 133
Query 117 NIAGDSPSCRNLVLDGGALPPLLEQL--NTPGSKV----AMLRNATWTLSNLCRGKPA-P 169
+A P + L++D GALP L+ L N GS +++R A ++NL +
Sbjct 134 LLA-IKPEYQKLIVDKGALPHLVNLLKRNKDGSSSRAVNSVIRRAADAITNLAHENSSIK 192
Query 170 VFEKVSPALPALAVLIYSDDAEVLTDACWALSYLSDGPNDRIKAVLEAGVCARLVELLSH 229
+V +P L L+ D++V A AL L+ +D ++E L+ +L
Sbjct 193 TRVRVEGGIPPLVELLEFSDSKVQRAAAGALRTLAFKNDDNKNQIVECNALPTLILMLGS 252
Query 230 QSNLVQTPALRAVGNIVTGDDTQTQVVLTCGALPKLLTLLSSTKKNVQKEACWTVSNITA 289
+ + A+ +GN+V + VLT GAL ++ LLSS Q+EA + +
Sbjct 253 EDAAIHYEAVGVIGNLVHSSPHIKKEVLTAGALQPVIGLLSSCCPESQREAALLLGQFAS 312
Query 290 GNKDQIQEVLDSGAIPALLHLLETADFDTKKEAAWAV 326
+ D ++ GA+ L+ +L++ D K+ +A+A+
Sbjct 313 TDSDCKVHIVQRGAVRPLIEMLQSPDVQLKEMSAFAL 349
Score = 54.7 bits (130), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 58/254 (22%), Positives = 102/254 (40%), Gaps = 41/254 (16%)
Query 109 EQAVWALGNIAGDSPSCRNLVLDGGALPPLLEQLNTPGSKVAMLRNATWTLSNLCRGKPA 168
++A L +A ++ N+++DGGA+P L+ L P + +L KP
Sbjct 72 KRATQVLAELAKNAEDLVNVIVDGGAVPALMTHLQAP----------PYNDGDLAE-KPY 120
Query 169 PVFEKVSPALPALAVLIYSDDAEVLTDACWALSYLSDGPNDRIKAVLEAGVCARLVELL- 227
+ EV + +AL L+ P + K +++ G LV LL
Sbjct 121 --------------------EHEVEKGSAFALGLLAIKPEYQ-KLIVDKGALPHLVNLLK 159
Query 228 -------SHQSNLVQTPALRAVGNIVTGDDTQTQVVLTCGALPKLLTLLSSTKKNVQKEA 280
S N V A A+ N+ + + V G +P L+ LL + VQ+ A
Sbjct 160 RNKDGSSSRAVNSVIRRAADAITNLAHENSSIKTRVRVEGGIPPLVELLEFSDSKVQRAA 219
Query 281 CWTVSNITAGNKDQIQEVLDSGAIPALLHLLETADFDTKKEAAWAVSNAVSGGTDMQVAL 340
+ + N D ++++ A+P L+ +L + D EA + N V ++ +
Sbjct 220 AGALRTLAFKNDDNKNQIVECNALPTLILMLGSEDAAIHYEAVGVIGNLVHSSPHIKKEV 279
Query 341 LVEKGCIPPLCSLL 354
L G + P+ LL
Sbjct 280 LT-AGALQPVIGLL 292
Score = 53.5 bits (127), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 79/280 (28%), Positives = 124/280 (44%), Gaps = 17/280 (6%)
Query 39 PPIQE-VIDAGAVPHFVRFL-------SMHGRYQLQFEAAWALTNIAS-GTQEQTRVVVE 89
P Q+ ++D GA+PH V L S + AA A+TN+A + +TRV VE
Sbjct 139 PEYQKLIVDKGALPHLVNLLKRNKDGSSSRAVNSVIRRAADAITNLAHENSSIKTRVRVE 198
Query 90 CGAIPLFVELLRCPKEEVREQAVWALGNIAGDSPSCRNLVLDGGALPPLLEQLNTPGSKV 149
G IP VELL +V+ A AL +A + +N +++ ALP L+ L + +
Sbjct 199 -GGIPPLVELLEFSDSKVQRAAAGALRTLAFKNDDNKNQIVECNALPTLILMLGSEDA-- 255
Query 150 AMLRNATWTLSNLCRGKPAPVFEKVSP-ALPALAVLIYSDDAEVLTDACWALSYLSDGPN 208
A+ A + NL P E ++ AL + L+ S E +A L + +
Sbjct 256 AIHYEAVGVIGNLVHSSPHIKKEVLTAGALQPVIGLLSSCCPESQREAALLLGQFASTDS 315
Query 209 DRIKAVLEAGVCARLVELLSHQSNLVQTPALRAVGNIVTGDDTQTQV-VLTCGALPKLLT 267
D +++ G L+E+L QS VQ + A D Q + G L LL
Sbjct 316 DCKVHIVQRGAVRPLIEML--QSPDVQLKEMSAFALGRLAQDAHNQAGIAHSGGLGPLLK 373
Query 268 LLSSTKKNVQKEACWTVSNITAGNKDQIQEVLDSGAIPAL 307
LL S ++Q A + + + A N+D + + + G I L
Sbjct 374 LLDSRNGSLQHNAAFALYGL-ADNEDNVSDFIRVGGIQKL 412
Score = 50.1 bits (118), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 33/112 (29%), Positives = 53/112 (47%), Gaps = 1/112 (0%)
Query 33 LSLENNPPIQEVIDAGAVPHFVRFLSMHGRYQLQFEAAWALTNIASGTQEQTRVVVECGA 92
L+ EN+ V G +P V L ++Q AA AL +A + +VEC A
Sbjct 184 LAHENSSIKTRVRVEGGIPPLVELLEFSDS-KVQRAAAGALRTLAFKNDDNKNQIVECNA 242
Query 93 IPLFVELLRCPKEEVREQAVWALGNIAGDSPSCRNLVLDGGALPPLLEQLNT 144
+P + +L + +AV +GN+ SP + VL GAL P++ L++
Sbjct 243 LPTLILMLGSEDAAIHYEAVGVIGNLVHSSPHIKKEVLTAGALQPVIGLLSS 294
Score = 35.0 bits (79), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 34/142 (23%), Positives = 59/142 (41%), Gaps = 22/142 (15%)
Query 278 KEACWTVSNITAGNKDQIQEVLDSGAIPALLHLLETADFDTKKEAAWAVSNAVSGGTDMQ 337
K A ++ + +D + ++D GA+PAL+ L+ ++ A + V G+
Sbjct 72 KRATQVLAELAKNAEDLVNVIVDGGAVPALMTHLQAPPYNDGDLAEKPYEHEVEKGSAFA 131
Query 338 VALL----------VEKGCIPPLCSLLGCMD--------NKIIMVALEALNAILRTGKKI 379
+ LL V+KG +P L +LL N +I A +A+ + I
Sbjct 132 LGLLAIKPEYQKLIVDKGALPHLVNLLKRNKDGSSSRAVNSVIRRAADAITNLAHENSSI 191
Query 380 K---ETEGLSVNPYATLIEQAD 398
K EG + P L+E +D
Sbjct 192 KTRVRVEG-GIPPLVELLEFSD 212
> Hs18562577
Length=548
Score = 83.6 bits (205), Expect = 7e-16, Method: Compositional matrix adjust.
Identities = 53/132 (40%), Positives = 74/132 (56%), Gaps = 16/132 (12%)
Query 181 LAVLIY---SDDAEVLT-DACWALSYLSDGPNDRIKAVLEAGVCARLVELLSHQSNLVQT 236
LA+L++ DD L +A WAL+ ++ G +++ +AV H VQT
Sbjct 141 LAILVHCLERDDNPCLQFEAAWALTNIAFGTSEQTQAVAH------------HHEVKVQT 188
Query 237 PALRAVGNIVTGDDTQTQVVLTCGALPKLLTLLSSTKKNVQKEACWTVSNITAGNKDQIQ 296
ALRAVGNI+ G D QTQVVL C AL LL+ K+ + KEA W +SNITAG++ Q
Sbjct 189 AALRAVGNIIMGTDEQTQVVLNCDALLHFPALLTHPKEKINKEAVWFLSNITAGSQQQSL 248
Query 297 EVLDSGAIPALL 308
LDS + ++
Sbjct 249 GRLDSQRVASIY 260
Score = 72.4 bits (176), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 51/155 (32%), Positives = 70/155 (45%), Gaps = 30/155 (19%)
Query 27 QLFRRALSLENNPPIQEVIDAGAVPHFVRFLSMHGRYQLQFEAAWALTNIA--------- 77
Q R+ LS + NPPI ++I +G + V L LQFEAAWALTNIA
Sbjct 118 QAARKLLSSDRNPPIDDLIKSGILAILVHCLERDDNPCLQFEAAWALTNIAFGTSEQTQA 177
Query 78 ---------------------SGTQEQTRVVVECGAIPLFVELLRCPKEEVREQAVWALG 116
GT EQT+VV+ C A+ F LL PKE++ ++AVW L
Sbjct 178 VAHHHEVKVQTAALRAVGNIIMGTDEQTQVVLNCDALLHFPALLTHPKEKINKEAVWFLS 237
Query 117 NIAGDSPSCRNLVLDGGALPPLLEQLNTPGSKVAM 151
NI S LD + + GS++++
Sbjct 238 NITAGSQQQSLGRLDSQRVASIYAGAGGSGSQISL 272
Score = 42.7 bits (99), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 43/144 (29%), Positives = 57/144 (39%), Gaps = 13/144 (9%)
Query 199 ALSYLSDGPNDRIKAVLEAGVCARLVELLSHQSN-LVQTPALRAVGNIVTGDDTQTQVVL 257
A LS N I ++++G+ A LV L N +Q A A+ NI G QTQ V
Sbjct 120 ARKLLSSDRNPPIDDLIKSGILAILVHCLERDDNPCLQFEAAWALTNIAFGTSEQTQAVA 179
Query 258 TCGALPKLLTLLSSTKKNVQKEACWTVSNITAGNKDQIQEVLDSGAIPALLHLLETADFD 317
+ VQ A V NI G +Q Q VL+ A+ LL
Sbjct 180 HHHEVK------------VQTAALRAVGNIIMGTDEQTQVVLNCDALLHFPALLTHPKEK 227
Query 318 TKKEAAWAVSNAVSGGTDMQVALL 341
KEA W +SN +G + L
Sbjct 228 INKEAVWFLSNITAGSQQQSLGRL 251
> At1g32880
Length=183
Score = 81.3 bits (199), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 46/122 (37%), Positives = 70/122 (57%), Gaps = 2/122 (1%)
Query 294 QIQEVLDSGAIPALLHLLETADFDTKKEAAWAVSNAVSGGTDMQVALLVEKGCIPPLCSL 353
++Q V+D+ IP L+ L + A+FD KKE+ A+SNA G+ Q+ +VE+ CI PLC +
Sbjct 40 KLQSVIDANLIPTLVKLTQNAEFDMKKESVCAISNATLLGSHDQIKYMVEQSCIKPLCDI 99
Query 354 LGCMDNKIIMVALEALNAILRTGKKIKETEGLSVNPYATLIEQADGATALEKLQDSTNEN 413
L C D K I+ L+ + L+ G+ K G V+ Y LIE A+G + LQ N
Sbjct 100 LFCPDVKTILKCLDGMENTLKVGEAEKNA-GDDVSWYTRLIE-AEGLDKILNLQRHENIE 157
Query 414 VF 415
++
Sbjct 158 IY 159
> CE06461
Length=524
Score = 80.1 bits (196), Expect = 9e-15, Method: Compositional matrix adjust.
Identities = 67/297 (22%), Positives = 142/297 (47%), Gaps = 12/297 (4%)
Query 122 SPSCRNLVLDGGALPPLLEQLNTPGSKVAMLRNATWTLSNLCR--GKPAPVFEKVSPALP 179
S + RNL LD + L++ +T + + R+ W + C + +P ++++P L
Sbjct 185 SITYRNLALDCAIVSELIDA-STINMSIILHRSLMWLVFLFCEKLDRCSPHVDEIAPLLE 243
Query 180 ALAVLIYSDDAEVLTDACWALSYLSDGPNDRIKAVLEAGVCARLVELLSHQSNLVQTPAL 239
++ I S DA V TDA + + L++ P + + +C++LV L + +
Sbjct 244 IISNGIQSTDAMVQTDAASSCASLAEWP-PIYHYMSDLKLCSKLVANLRNDKGNARPKVK 302
Query 240 RAVGNIVTGDDTQTQVVLTCGALPKLLTLLSSTKKNVQKEACWTVSNITAGNKDQIQEVL 299
+ +I+ T+ ++ G L L ++ + + +E C+ +SNI + I +++
Sbjct 303 AGINSIIQATGYFTEEMIDAGLLEVLKGFVNVSY--MSQEVCFIISNICVEGEQTIDKLI 360
Query 300 DSGAIPALLHLLETADFDTKKEAAWAVSNAVSGGTDMQVALLVEKGCIPPLCSLLGCMDN 359
SG + + ++E +++ +++EAA+ + + + + +VE G + LL CMD
Sbjct 361 SSGVLREVARVMEASEYRSRREAAFVICHCCASANQKHLEYVVELGMLSAFTDLLTCMDV 420
Query 360 KIIMVALEALNAILRTGKKIKETEGL--SVNPYATLIEQADGATALEKLQDSTNENV 414
++ L+A+ +L+ G E L + NP A +E+ LE L +S + ++
Sbjct 421 SLVSYILDAIYLLLQFG----EMRLLPDNSNPVAIKLEEIGCREKLEFLCESQSVDI 473
Score = 32.3 bits (72), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 20/71 (28%), Positives = 35/71 (49%), Gaps = 2/71 (2%)
Query 33 LSLENNPPIQEVIDAGAVPHFVRFLSMHGRYQLQFEAAWALTNI-ASGTQEQTRVVVECG 91
+ +E I ++I +G + R + Y+ + EAA+ + + AS Q+ VVE G
Sbjct 348 ICVEGEQTIDKLISSGVLREVARVMEA-SEYRSRREAAFVICHCCASANQKHLEYVVELG 406
Query 92 AIPLFVELLRC 102
+ F +LL C
Sbjct 407 MLSAFTDLLTC 417
> Hs6912678
Length=509
Score = 73.6 bits (179), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 73/292 (25%), Positives = 132/292 (45%), Gaps = 5/292 (1%)
Query 37 NNPPIQEVIDAGAVPHFVRFLSMHGRYQLQFEAAWALTNIASGTQEQTRVVVECGAIPLF 96
N+ + V+ +P V L+ R+ + AA+ L + + + + +V+CGA+
Sbjct 72 NDDLAEAVVKCDILPQLVYSLAEQNRF-YKKAAAFVLRAVGKHSPQLAQAIVDCGALDTL 130
Query 97 VELLRCPKEEVREQAVWALGNIAGDSPSCRNLVLDGGALPPLLEQLNTPGSKVAMLRNAT 156
V L V+E A WAL IA + V+D GA+P L+ + P ++A+ R A
Sbjct 131 VICLEDFDPGVKEAAAWALRYIARHNAELSQAVVDAGAVPLLVLCIQEP--EIALKRIAA 188
Query 157 WTLSNLCRGKPAPVFEKVSP-ALPALAVLIYSDDAEVLTDACWALSYLSDGPNDRIKAVL 215
LS++ + P V A+ LA +I + DA++ ALS +S D + V+
Sbjct 189 SALSDIAKHSPELAQTVVDAGAVAHLAQMILNPDAKLKHQILSALSQVSKHSVDLAEMVV 248
Query 216 EAGVCARLVELLSHQSNLVQTPALRAVGNIVTGDDTQTQVVLTCGALPKLLTLLSSTKKN 275
EA + ++ L + V+ A + I +Q+V+ G + ++ + S K N
Sbjct 249 EAEIFPVVLTCLKDKDEYVKKNASTLIREIAKHTPELSQLVVNAGGVAAVIDCIGSCKGN 308
Query 276 VQKEACWTVSNITAGNKDQIQEVLDSGAIPAL-LHLLETADFDTKKEAAWAV 326
+ + + A +++ V+ S +P L + L E + K AAWA+
Sbjct 309 TRLPGIMMLGYVAAHSENLAMAVIISKGVPQLSVCLSEEPEDHIKAAAAWAL 360
Score = 66.2 bits (160), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 76/340 (22%), Positives = 148/340 (43%), Gaps = 24/340 (7%)
Query 7 LQQLVQALASGVPDAELEGTQLFRRALSL------ENNPPI-QEVIDAGAVPHFVRFLSM 59
L QLV +LA E + +++A + +++P + Q ++D GA+ V L
Sbjct 85 LPQLVYSLA--------EQNRFYKKAAAFVLRAVGKHSPQLAQAIVDCGALDTLVICLED 136
Query 60 HGRYQLQFEAAWALTNIASGTQEQTRVVVECGAIPLFVELLRCPKEEVREQAVWALGNIA 119
++ AAWAL IA E ++ VV+ GA+PL V ++ P+ ++ A AL +IA
Sbjct 137 FDP-GVKEAAAWALRYIARHNAELSQAVVDAGAVPLLVLCIQEPEIALKRIAASALSDIA 195
Query 120 GDSPSCRNLVLDGGALPPLLEQLNTPGSKVAMLRNATWTLSNLCRGKPAPVFEKVSPA-- 177
SP V+D GA+ L + + P +K L++ + + + E V A
Sbjct 196 KHSPELAQTVVDAGAVAHLAQMILNPDAK---LKHQILSALSQVSKHSVDLAEMVVEAEI 252
Query 178 LPALAVLIYSDDAEVLTDACWALSYLSDGPNDRIKAVLEAGVCARLVELLSHQSNLVQTP 237
P + + D V +A + ++ + + V+ AG A +++ + + P
Sbjct 253 FPVVLTCLKDKDEYVKKNASTLIREIAKHTPELSQLVVNAGGVAAVIDCIGSCKGNTRLP 312
Query 238 ALRAVGNIVTGDDTQTQVVLTCGALPKLLTLLSST-KKNVQKEACWTVSNITAGNKDQIQ 296
+ +G + + V+ +P+L LS + +++ A W + I + +
Sbjct 313 GIMMLGYVAAHSENLAMAVIISKGVPQLSVCLSEEPEDHIKAAAAWALGQIGRHTPEHAR 372
Query 297 EVLDSGAIPALLHLLETADF--DTKKEAAWAVSNAVSGGT 334
V + +P LL L + + D + ++ A+ N + T
Sbjct 373 AVAVTNTLPVLLSLYMSTESSEDLQVKSKKAIKNILQKCT 412
Score = 63.9 bits (154), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 75/330 (22%), Positives = 141/330 (42%), Gaps = 12/330 (3%)
Query 26 TQLFRRALSLENNPPIQEVIDAGAVPHFVRFLSMHGRYQLQFEAAWALTNIASGTQEQTR 85
TQ + L P E + V +R L + +Q AA AL +A+ +
Sbjct 18 TQFVQMVAELATRPQNIETLQNAGVMSLLRTLLLDVVPTIQQTAALALGRLANYNDDLAE 77
Query 86 VVVECGAIPLFVELLRCPKEEVREQAVWALGNIAGDSPSCRNLVLDGGALPPL---LEQL 142
VV+C +P V L ++ A + L + SP ++D GAL L LE
Sbjct 78 AVVKCDILPQLVYSLAEQNRFYKKAAAFVLRAVGKHSPQLAQAIVDCGALDTLVICLEDF 137
Query 143 NTPGSKVAMLRNATWTLSNLCRGKPAPVFEKV--SPALPALAVLIYSDDAEVLTDACWAL 200
+ PG K A A W L + R A + + V + A+P L + I + + A AL
Sbjct 138 D-PGVKEA----AAWALRYIARHN-AELSQAVVDAGAVPLLVLCIQEPEIALKRIAASAL 191
Query 201 SYLSDGPNDRIKAVLEAGVCARLVELLSHQSNLVQTPALRAVGNIVTGDDTQTQVVLTCG 260
S ++ + + V++AG A L +++ + ++ L A+ + ++V+
Sbjct 192 SDIAKHSPELAQTVVDAGAVAHLAQMILNPDAKLKHQILSALSQVSKHSVDLAEMVVEAE 251
Query 261 ALPKLLTLLSSTKKNVQKEACWTVSNITAGNKDQIQEVLDSGAIPALLHLLETADFDTKK 320
P +LT L + V+K A + I + Q V+++G + A++ + + +T+
Sbjct 252 IFPVVLTCLKDKDEYVKKNASTLIREIAKHTPELSQLVVNAGGVAAVIDCIGSCKGNTRL 311
Query 321 EAAWAVSNAVSGGTDMQVALLVEKGCIPPL 350
+ + ++ +A+++ KG +P L
Sbjct 312 PGIMMLGYVAAHSENLAMAVIISKG-VPQL 340
Score = 59.3 bits (142), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 50/189 (26%), Positives = 90/189 (47%), Gaps = 7/189 (3%)
Query 196 ACWALSYLSDGPNDRIKAVLEAGVCARLVELLSHQSNLVQTPA---LRAVGNIVTGDDTQ 252
A AL L++ +D +AV++ + +LV L+ Q+ + A LRAVG
Sbjct 61 AALALGRLANYNDDLAEAVVKCDILPQLVYSLAEQNRFYKKAAAFVLRAVGK---HSPQL 117
Query 253 TQVVLTCGALPKLLTLLSSTKKNVQKEACWTVSNITAGNKDQIQEVLDSGAIPALLHLLE 312
Q ++ CGAL L+ L V++ A W + I N + Q V+D+GA+P L+ ++
Sbjct 118 AQAIVDCGALDTLVICLEDFDPGVKEAAAWALRYIARHNAELSQAVVDAGAVPLLVLCIQ 177
Query 313 TADFDTKKEAAWAVSNAVSGGTDMQVALLVEKGCIPPLCSLLGCMDNKIIMVALEALNAI 372
+ K+ AA A+S+ ++ +V+ G + L ++ D K+ L AL+ +
Sbjct 178 EPEIALKRIAASALSDIAKHSPEL-AQTVVDAGAVAHLAQMILNPDAKLKHQILSALSQV 236
Query 373 LRTGKKIKE 381
+ + E
Sbjct 237 SKHSVDLAE 245
Score = 52.4 bits (124), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 43/172 (25%), Positives = 81/172 (47%), Gaps = 1/172 (0%)
Query 211 IKAVLEAGVCARLVELLSHQSNLVQTPALRAVGNIVTGDDTQTQVVLTCGALPKLLTLLS 270
I+ + AGV + L LL +Q A A+G + +D + V+ C LP+L+ L+
Sbjct 34 IETLQNAGVMSLLRTLLLDVVPTIQQTAALALGRLANYNDDLAEAVVKCDILPQLVYSLA 93
Query 271 STKKNVQKEACWTVSNITAGNKDQIQEVLDSGAIPALLHLLETADFDTKKEAAWAVSNAV 330
+ +K A + + + + Q ++D GA+ L+ LE D K+ AAWA+
Sbjct 94 EQNRFYKKAAAFVLRAVGKHSPQLAQAIVDCGALDTLVICLEDFDPGVKEAAAWALRYIA 153
Query 331 SGGTDMQVALLVEKGCIPPLCSLLGCMDNKIIMVALEALNAILRTGKKIKET 382
++ A +V+ G +P L + + + +A AL+ I + ++ +T
Sbjct 154 RHNAELSQA-VVDAGAVPLLVLCIQEPEIALKRIAASALSDIAKHSPELAQT 204
> YEL013w
Length=578
Score = 56.6 bits (135), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 67/253 (26%), Positives = 118/253 (46%), Gaps = 7/253 (2%)
Query 64 QLQFEAAWALTNIASGTQEQTRVVVECGAIPLFVELLRCPKEEVREQAVWALGNIAGDSP 123
Q+Q A AL N+A E ++VE G + + + EV+ AV + N+A
Sbjct 100 QIQVAACAALGNLAVNN-ENKLLIVEMGGLEPLINQMMGDNVEVQCNAVGCITNLATRDD 158
Query 124 SCRNLVLDGGALPPLLEQLNTPGSKVAMLRNATWTLSNLCRGKPAPVFEKVSPALPALAV 183
+ ++ + GAL PL + + +V RNAT L N+ + + A+P L
Sbjct 159 N-KHKIATSGALIPLTKLAKSKHIRVQ--RNATGALLNMTHSEENRKELVNAGAVPVLVS 215
Query 184 LIYSDDAEVLTDACWALSYLS-DGPNDRIKAVLEAGVCARLVELLSHQSNLVQTPALRAV 242
L+ S D +V ALS ++ D N + A E + ++LV L+ S+ V+ A A+
Sbjct 216 LLSSTDPDVQYYCTTALSNIAVDEANRKKLAQTEPRLVSKLVSLMDSPSSRVKCQATLAL 275
Query 243 GNIVTGDDTQTQVVLTCGALPKLLTLLSSTKKNVQKEACWTVSNITAGNKDQIQEVLDSG 302
N+ + Q ++V G LP L+ L+ S + + + NI+ ++ ++D+G
Sbjct 276 RNLASDTSYQLEIV-RAGGLPHLVKLIQSDSIPLVLASVACIRNISIHPLNE-GLIVDAG 333
Query 303 AIPALLHLLETAD 315
+ L+ LL+ D
Sbjct 334 FLKPLVRLLDYKD 346
Score = 55.5 bits (132), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 47/177 (26%), Positives = 87/177 (49%), Gaps = 3/177 (1%)
Query 178 LPALAVLIYSDDAEVLTDACWALSYLSDGPNDRIKAVLEAGVCARLVELLSHQSNLVQTP 237
L + +L+ S D ++ AC AL L+ +++ ++E G L+ + + VQ
Sbjct 87 LEPILILLQSQDPQIQVAACAALGNLAVNNENKL-LIVEMGGLEPLINQMMGDNVEVQCN 145
Query 238 ALRAVGNIVTGDDTQTQVVLTCGALPKLLTLLSSTKKNVQKEACWTVSNITAGNKDQIQE 297
A+ + N+ T DD + ++ T GAL L L S VQ+ A + N+T +++ +E
Sbjct 146 AVGCITNLATRDDNKHKIA-TSGALIPLTKLAKSKHIRVQRNATGALLNMTHSEENR-KE 203
Query 298 VLDSGAIPALLHLLETADFDTKKEAAWAVSNAVSGGTDMQVALLVEKGCIPPLCSLL 354
++++GA+P L+ LL + D D + A+SN + + E + L SL+
Sbjct 204 LVNAGAVPVLVSLLSSTDPDVQYYCTTALSNIAVDEANRKKLAQTEPRLVSKLVSLM 260
Score = 55.1 bits (131), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 60/233 (25%), Positives = 107/233 (45%), Gaps = 10/233 (4%)
Query 57 LSMHGRYQLQFEAAWALTNIASGTQEQTRVVVECGAIPLFVELLRCPKEEVREQAVWALG 116
L+ ++Q A AL N+ + ++E + +V GA+P+ V LL +V+ AL
Sbjct 175 LAKSKHIRVQRNATGALLNM-THSEENRKELVNAGAVPVLVSLLSSTDPDVQYYCTTALS 233
Query 117 NIAGDSPSCRNLV-LDGGALPPLLEQLNTPGSKVAMLRNATWTLSNLCRGKPAPVFEKVS 175
NIA D + + L + + L+ +++P S+V AT L NL + +
Sbjct 234 NIAVDEANRKKLAQTEPRLVSKLVSLMDSPSSRVKC--QATLALRNLASDTSYQLEIVRA 291
Query 176 PALPALAVLIYSDDAEVLTDACWALSYLSDGP-NDRIKAVLEAGVCARLVELLSHQ-SNL 233
LP L LI SD ++ + + +S P N+ + +++AG LV LL ++ S
Sbjct 292 GGLPHLVKLIQSDSIPLVLASVACIRNISIHPLNEGL--IVDAGFLKPLVRLLDYKDSEE 349
Query 234 VQTPALRAVGNIVTGDDTQTQVVLTCGALPKLLTLLSSTKKNVQKE--ACWTV 284
+Q A+ + N+ + + GA+ K L + +VQ E AC+ +
Sbjct 350 IQCHAVSTLRNLAASSEKNRKEFFESGAVEKCKELALDSPVSVQSEISACFAI 402
Score = 48.5 bits (114), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 56/222 (25%), Positives = 99/222 (44%), Gaps = 13/222 (5%)
Query 178 LPALAVLIYSDDAEVLTDACWALSYLSDGPNDRI-KAVLEAGVCARLVELLSHQSNLVQT 236
L AL L+YSD+ + A A + +++ ++ + VLE ++ LL Q +Q
Sbjct 49 LKALTTLVYSDNLNLQRSAALAFAEITEKYVRQVSREVLEP-----ILILLQSQDPQIQV 103
Query 237 PALRAVGNIVTGDDTQTQVVLTCGALPKLLTLLSSTKKNVQKEACWTVSNITAGNKDQIQ 296
A A+GN+ ++ + +V G L L+ + VQ A ++N+ A D
Sbjct 104 AACAALGNLAVNNENKLLIV-EMGGLEPLINQMMGDNVEVQCNAVGCITNL-ATRDDNKH 161
Query 297 EVLDSGAIPALLHLLETADFDTKKEAAWAVSNAVSGGTDMQVALLVEKGCIPPLCSLLGC 356
++ SGA+ L L ++ ++ A A+ N + + LV G +P L SLL
Sbjct 162 KIATSGALIPLTKLAKSKHIRVQRNATGALLNMTHSEENRKE--LVNAGAVPVLVSLLSS 219
Query 357 MDNKIIMVALEALNAIL---RTGKKIKETEGLSVNPYATLIE 395
D + AL+ I KK+ +TE V+ +L++
Sbjct 220 TDPDVQYYCTTALSNIAVDEANRKKLAQTEPRLVSKLVSLMD 261
Score = 48.1 bits (113), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 52/191 (27%), Positives = 82/191 (42%), Gaps = 14/191 (7%)
Query 42 QEVIDAGAVPHFVRFLSMHGRYQLQFEAAWALTNIASGTQEQTRVV-VECGAIPLFVELL 100
+E+++AGAVP V LS +Q+ AL+NIA + ++ E + V L+
Sbjct 202 KELVNAGAVPVLVSLLSSTDP-DVQYYCTTALSNIAVDEANRKKLAQTEPRLVSKLVSLM 260
Query 101 RCPKEEVREQAVWALGNIAGDSPSCRNLVLDGGALPPLLEQLNTPG-----SKVAMLRNA 155
P V+ QA AL N+A D+ +V GG LP L++ + + + VA +RN
Sbjct 261 DSPSSRVKCQATLALRNLASDTSYQLEIVRAGG-LPHLVKLIQSDSIPLVLASVACIRNI 319
Query 156 TWTLSNLCRGKPAPVFEKVSPALPALAVLIYSDDAEVLTDACWALSYLSDGPNDRIKAVL 215
+ N A + P + +L Y D E+ A L L+ K
Sbjct 320 SIHPLNEGLIVDAGFLK------PLVRLLDYKDSEEIQCHAVSTLRNLAASSEKNRKEFF 373
Query 216 EAGVCARLVEL 226
E+G + EL
Sbjct 374 ESGAVEKCKEL 384
Score = 42.0 bits (97), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 62/253 (24%), Positives = 113/253 (44%), Gaps = 17/253 (6%)
Query 59 MHGRYQLQFEAAWALTNIASGTQEQTRVVVECGAIPLFVELLRCPKEEVREQAVWALGNI 118
M ++Q A +TN+A+ + ++ IPL +L + V+ A AL N+
Sbjct 136 MGDNVEVQCNAVGCITNLATRDDNKHKIATSGALIPL-TKLAKSKHIRVQRNATGALLNM 194
Query 119 AGDSPSCRNLVLDGGALPPLLEQLNTPGSKVAMLRNATWTLSNLC-----RGKPAPVFEK 173
+ + LV + GA+P L+ L++ V T LSN+ R K A +
Sbjct 195 THSEENRKELV-NAGAVPVLVSLLSSTDPDVQYY--CTTALSNIAVDEANRKKLA----Q 247
Query 174 VSPALPA-LAVLIYSDDAEVLTDACWALSYLSDGPNDRIKAVLEAGVCARLVELLSHQSN 232
P L + L L+ S + V A AL L+ + +++ ++ AG LV+L+ S
Sbjct 248 TEPRLVSKLVSLMDSPSSRVKCQATLALRNLASDTSYQLE-IVRAGGLPHLVKLIQSDSI 306
Query 233 LVQTPALRAVGNIVTGDDTQTQVVLTCGALPKLLTLLS-STKKNVQKEACWTVSNITAGN 291
+ ++ + NI + +++ G L L+ LL + +Q A T+ N+ A +
Sbjct 307 PLVLASVACIRNI-SIHPLNEGLIVDAGFLKPLVRLLDYKDSEEIQCHAVSTLRNLAASS 365
Query 292 KDQIQEVLDSGAI 304
+ +E +SGA+
Sbjct 366 EKNRKEFFESGAV 378
Score = 33.1 bits (74), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 18/62 (29%), Positives = 31/62 (50%), Gaps = 2/62 (3%)
Query 40 PIQE--VIDAGAVPHFVRFLSMHGRYQLQFEAAWALTNIASGTQEQTRVVVECGAIPLFV 97
P+ E ++DAG + VR L ++Q A L N+A+ +++ + E GA+
Sbjct 323 PLNEGLIVDAGFLKPLVRLLDYKDSEEIQCHAVSTLRNLAASSEKNRKEFFESGAVEKCK 382
Query 98 EL 99
EL
Sbjct 383 EL 384
> At3g46510
Length=660
Score = 53.9 bits (128), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 69/267 (25%), Positives = 117/267 (43%), Gaps = 11/267 (4%)
Query 60 HGRYQLQFEAAWALTNIASGTQEQTRVVVECGAIPLFVELLRCPKEEVREQAVWALGNIA 119
+G + Q AA + +A + + E GAIPL V LL P ++E +V AL N++
Sbjct 363 YGNPEDQRSAAGEIRLLAKRNADNRVAIAEAGAIPLLVGLLSTPDSRIQEHSVTALLNLS 422
Query 120 GDSPSCRN---LVLDGGALPPLLEQLNTPGSKVAMLRNATWTLSNLCRGKPAPVFEKVSP 176
C N ++ GA+P +++ L GS A NA TL +L V
Sbjct 423 ----ICENNKGAIVSAGAIPGIVQVLKK-GSMEAR-ENAAATLFSLSVIDENKVTIGALG 476
Query 177 ALPALAVLIYSDDAEVLTDACWALSYLSDGPNDRIKAVLEAGVCARLVELLSHQSNLVQT 236
A+P L VL+ DA AL L ++ KA+ AGV L LL+ + +
Sbjct 477 AIPPLVVLLNEGTQRGKKDAATALFNLCIYQGNKGKAI-RAGVIPTLTRLLTEPGSGMVD 535
Query 237 PALRAVGNIVTGDDTQTQVVLTCGALPKLLTLLSSTKKNVQKEACWTVSNITAGNKDQIQ 296
AL A+ I++ ++ + A+P L+ + + ++ A + ++ +G+ +
Sbjct 536 EAL-AILAILSSHPEGKAIIGSSDAVPSLVEFIRTGSPRNRENAAAVLVHLCSGDPQHLV 594
Query 297 EVLDSGAIPALLHLLETADFDTKKEAA 323
E G + L+ L K++AA
Sbjct 595 EAQKLGLMGPLIDLAGNGTDRGKRKAA 621
Score = 42.7 bits (99), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 49/199 (24%), Positives = 92/199 (46%), Gaps = 26/199 (13%)
Query 189 DAEVLTDACWALSY---------------LSDGPNDRIKAVLEAGVCARLVELLSHQSNL 233
+A + D W L+Y L+ D A+ EAG LV LLS +
Sbjct 350 EANKIEDLMWRLAYGNPEDQRSAAGEIRLLAKRNADNRVAIAEAGAIPLLVGLLSTPDSR 409
Query 234 VQTPALRAVGNIVTGDDTQTQVVLTCGALPKLLTLLSSTKKNVQKEACWTVSNITAGNKD 293
+Q ++ A+ N+ ++ + +V + GA+P ++ +L ++ A T+ +++ +++
Sbjct 410 IQEHSVTALLNLSICENNKGAIV-SAGAIPGIVQVLKKGSMEARENAAATLFSLSVIDEN 468
Query 294 QIQEVLDSGAIPALLHLLETADFDTKKEAAWAVSN-AVSGGTDMQVALLVEKGCIPPLCS 352
++ + GAIP L+ LL KK+AA A+ N + G + + G IP L
Sbjct 469 KVT-IGALGAIPPLVVLLNEGTQRGKKDAATALFNLCIYQGNKGKA---IRAGVIPTLTR 524
Query 353 LL-----GCMDNKIIMVAL 366
LL G +D + ++A+
Sbjct 525 LLTEPGSGMVDEALAILAI 543
> At1g77460
Length=2110
Score = 51.2 bits (121), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 63/236 (26%), Positives = 104/236 (44%), Gaps = 10/236 (4%)
Query 108 REQAVWALGNIAGDSPSCRNLVLDGGALPPLLEQLNTPGSKVAMLRNATWTLSNLCRGKP 167
+E L IA R L+ G PL + G+ +A + N L LC+ K
Sbjct 30 KELTTARLLGIAKGKREARRLIGSYGQAMPLFISMLRNGTTLAKV-NVASILCVLCKDKD 88
Query 168 APVFEKVSPALPALAVLIYSDDAEVLTDACWALSYLSDG--PNDRI--KAVLEAGVCARL 223
+ + +P L ++ S E A A+ +S ND I K + GV L
Sbjct 89 LRLKVLLGGCIPPLLSVLKSGTMETRKAAAEAIYEVSSAGISNDHIGMKIFITEGVVPTL 148
Query 224 VELLS---HQSNLVQTPALRAVGNIVTGDDTQTQVVLTCGALPKLLTLLSSTKKNVQKEA 280
+ LS +Q +V+ A+ N+ DD ++ L + +++LLSS N Q A
Sbjct 149 WDQLSLKGNQDKVVEGYVTGALRNLCGVDDGYWRLTLEGSGVDIVVSLLSSDNPNSQANA 208
Query 281 CWTVSNITAGNKDQIQEVLDSGAIPALLHLLETA-DFDTKKEAAWAVSNAVSGGTD 335
++ + D IQ++L+SG + +L+ LLE D + + AA A+ A+S +D
Sbjct 209 ASLLARLVLSFCDSIQKILNSGVVKSLIQLLEQKNDINVRASAADAL-EALSANSD 263
> At5g58680
Length=357
Score = 49.3 bits (116), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 43/150 (28%), Positives = 78/150 (52%), Gaps = 5/150 (3%)
Query 205 DGPNDRIKAVLEAGVCARLVELLSHQSNLVQTPALRAVGNIVTGDDTQTQVVLTCGALPK 264
+ P +RIK + +AG LV L+S +Q + AV N+ D+ + +++++ GA+
Sbjct 91 NKPENRIK-LAKAGAIKPLVSLISSSDLQLQEYGVTAVLNLSLCDENK-EMIVSSGAVKP 148
Query 265 LLTLLSSTKKNVQKEACWTVSNITAGNKDQIQEVLDSGAIPALLHLLETADFDTKKEAAW 324
L+ L ++ A + ++ +++I + SGAIP L++LLE F KK+A+
Sbjct 149 LVNALRLGTPTTKENAACALLRLSQVEENKIT-IGRSGAIPLLVNLLENGGFRAKKDAST 207
Query 325 AVSNAVSGGTDMQVALLVEKGCIPPLCSLL 354
A+ + S + A VE G + PL L+
Sbjct 208 ALYSLCSTNENKTRA--VESGIMKPLVELM 235
Score = 39.7 bits (91), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 68/328 (20%), Positives = 132/328 (40%), Gaps = 54/328 (16%)
Query 50 VPHFVRFLSMHGRYQLQFEAAWALTNIASGTQEQTRVVVECGAIPLFVELLRCPKEEVRE 109
+ + + L + Q +AA + ++ E + + GAI V L+ +++E
Sbjct 62 IRNLITHLESSSSIEEQKQAAMEIRLLSKNKPENRIKLAKAGAIKPLVSLISSSDLQLQE 121
Query 110 QAVWALGNIAGDSPSCRNLVLDGGALPPLLE--QLNTPGSKVAMLRNATWTLSNLCRGKP 167
V A+ N++ + + +++ GA+ PL+ +L TP +K NA L L + +
Sbjct 122 YGVTAVLNLSLCDEN-KEMIVSSGAVKPLVNALRLGTPTTK----ENAACALLRLSQVEE 176
Query 168 APVFEKVSPALPALAVLIYSDDAEVLTDACWALSYLSDGPNDRIKAVLEAGVCARLVELL 227
+ S A+P L L+ + DA AL L ++ +AV E+G+ LVEL+
Sbjct 177 NKITIGRSGAIPLLVNLLENGGFRAKKDASTALYSLCSTNENKTRAV-ESGIMKPLVELM 235
Query 228 -SHQSNLVQTPALRAVGNIVTGDDTQTQVVLTCGALPKLLTLLSSTKKNVQKEACWTVSN 286
+S++V A V N
Sbjct 236 IDFESDMVDKSAF--------------------------------------------VMN 251
Query 287 ITAGNKDQIQEVLDSGAIPALLHLLETADFDTKKEAAWAVSNAVSGGTDMQVALLVEKGC 346
+ + V++ G +P L+ ++E A +KE + ++ + + + ++ +G
Sbjct 252 LLMSAPESKPAVVEEGGVPVLVEIVE-AGTQRQKEISVSILLQLCEESVVYRTMVAREGA 310
Query 347 IPPLCSLLGCMDNKIIMVALEALNAILR 374
+PPL +L ++ V EAL +LR
Sbjct 311 VPPLVALSQGSASRGAKVKAEALIELLR 338
> 7289855
Length=779
Score = 48.9 bits (115), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 33/136 (24%), Positives = 69/136 (50%), Gaps = 3/136 (2%)
Query 221 ARLVELLSHQSNLVQTPALRAVGNIVTGDDTQTQVVLTCGALPKLLTLLSSTKKNVQKEA 280
+ ++ LS+ S+ ++ A + ++ DD Q + G +P L+ LLS + K A
Sbjct 196 SEVISFLSNPSSAIKANAAAYLQHLCYMDDPNKQRTRSLGGIPPLVRLLSYDSPEIHKNA 255
Query 281 CWTVSNITAG--NKDQIQEVLDSGAIPALLHLLETADFDTKKEAAWAVSNAVSGGTDMQV 338
C + N++ G N + + + ++G I AL+HLL + KE V +S D++
Sbjct 256 CGALRNLSYGRQNDENKRGIKNAGGIAALVHLLCRSQETEVKELVTGVLWNMSSCEDLKR 315
Query 339 ALLVEKGCIPPLCSLL 354
+ ++++ + +CS++
Sbjct 316 S-IIDEALVAVVCSVI 330
> At3g54850
Length=639
Score = 48.5 bits (114), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 68/264 (25%), Positives = 117/264 (44%), Gaps = 5/264 (1%)
Query 60 HGRYQLQFEAAWALTNIASGTQEQTRVVVECGAIPLFVELLRCPKEEVREQAVWALGNIA 119
+G + Q AA L +A + + E GAIPL VELL P +E +V AL N++
Sbjct 363 NGTTEQQRAAAGELRLLAKRNVDNRVCIAEAGAIPLLVELLSSPDPRTQEHSVTALLNLS 422
Query 120 GDSPSCRNLVLDGGALPPLLEQLNTPGSKVAMLRNATWTLSNLCRGKPAPVFEKVSPALP 179
+ + + ++D GA+ ++E L GS A NA TL +L V + A+
Sbjct 423 INEGN-KGAIVDAGAITDIVEVLKN-GSMEAR-ENAAATLFSLSVIDENKVAIGAAGAIQ 479
Query 180 ALAVLIYSDDAEVLTDACWALSYLSDGPNDRIKAVLEAGVCARLVELLSHQSNLVQTPAL 239
AL L+ DA A+ L ++ +AV + G+ L LL + AL
Sbjct 480 ALISLLEEGTRRGKKDAATAIFNLCIYQGNKSRAV-KGGIVDPLTRLLKDAGGGMVDEAL 538
Query 240 RAVGNIVTGDDTQTQVVLTCGALPKLLTLLSSTKKNVQKEACWTVSNITAGNKDQIQEVL 299
+ + T + +T + ++P L+ ++ + ++ A + + GN +++
Sbjct 539 AILAILSTNQEGKTAIA-EAESIPVLVEIIRTGSPRNRENAAAILWYLCIGNIERLNVAR 597
Query 300 DSGAIPALLHLLETADFDTKKEAA 323
+ GA AL L E K++AA
Sbjct 598 EVGADVALKELTENGTDRAKRKAA 621
Score = 35.0 bits (79), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 48/180 (26%), Positives = 88/180 (48%), Gaps = 13/180 (7%)
Query 214 VLEAGVCARLVELLSHQSNLVQTPALRAVGNIVTGDDTQTQVVLTCGALPKLLTLLSSTK 273
+ EAG LVELLS Q ++ A+ N+ + + +V GA+ ++ +L +
Sbjct 390 IAEAGAIPLLVELLSSPDPRTQEHSVTALLNLSINEGNKGAIV-DAGAITDIVEVLKNGS 448
Query 274 KNVQKEACWTVSNITAGNKDQIQEVLDSGAIPALLHLLETADFDTKKEAAWAVSN-AVSG 332
++ A T+ +++ +++++ +GAI AL+ LLE KK+AA A+ N +
Sbjct 449 MEARENAAATLFSLSVIDENKVAIGA-AGAIQALISLLEEGTRRGKKDAATAIFNLCIYQ 507
Query 333 GTDMQVALLVEKGCIPPLCSLL-----GCMDNKIIMVALEALNAILRTGKKIKETEGLSV 387
G + V+ G + PL LL G +D + ++A+ + N +T I E E + V
Sbjct 508 GNKSRA---VKGGIVDPLTRLLKDAGGGMVDEALAILAILSTNQEGKTA--IAEAESIPV 562
Score = 32.0 bits (71), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 43/151 (28%), Positives = 66/151 (43%), Gaps = 10/151 (6%)
Query 260 GALPKLLTLLSSTKKNVQKEACWTVSN--ITAGNKDQIQEVLDSGAIPALLHLLETADFD 317
GA+P L+ LLSS Q+ + + N I GNK I +D+GAI ++ +L+ +
Sbjct 394 GAIPLLVELLSSPDPRTQEHSVTALLNLSINEGNKGAI---VDAGAITDIVEVLKNGSME 450
Query 318 TKKEAAWAVSNAVSGGTDMQVALLVEKGCIPPLCSLL--GCMDNKIIMVALEALNAILRT 375
++ AA + + D + G I L SLL G K A N +
Sbjct 451 ARENAAATLFSL--SVIDENKVAIGAAGAIQALISLLEEGTRRGKK-DAATAIFNLCIYQ 507
Query 376 GKKIKETEGLSVNPYATLIEQADGATALEKL 406
G K + +G V+P L++ A G E L
Sbjct 508 GNKSRAVKGGIVDPLTRLLKDAGGGMVDEAL 538
> At2g28830
Length=654
Score = 47.8 bits (112), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 49/187 (26%), Positives = 89/187 (47%), Gaps = 17/187 (9%)
Query 31 RALSLENNPPIQEVIDAGAVPHFVRFLSMHGRYQLQFEAAWALTNIASGTQEQTRVVVEC 90
R L+ +NN + +GA+P V L++ + Q A ++ N++ + + ++V
Sbjct 380 RLLAKQNNHNRVAIAASGAIPLLVNLLTISNDSRTQEHAVTSILNLSICQENKGKIVYSS 439
Query 91 GAIPLFVELLRCPKEEVREQAVWALGNIAGDSPSCRNLVLDG--GALPPLLEQLNTPGSK 148
GA+P V +L+ E RE A L ++ S N V G GA+PPL+ L + GS+
Sbjct 440 GAVPGIVHVLQKGSMEARENAAATLFSL---SVIDENKVTIGAAGAIPPLVTLL-SEGSQ 495
Query 149 VAMLRNATWTLSNLC-----RGKPAPVFEKVSPALPALAVLIYSDDAEVLTDACWALSYL 203
++A L NLC +GK + +P L L+ ++ ++ ++ L+ L
Sbjct 496 RGK-KDAATALFNLCIFQGNKGKAVR-----AGLVPVLMRLLTEPESGMVDESLSILAIL 549
Query 204 SDGPNDR 210
S P+ +
Sbjct 550 SSHPDGK 556
Score = 42.4 bits (98), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 27/100 (27%), Positives = 52/100 (52%), Gaps = 3/100 (3%)
Query 256 VLTCGALPKLLTLLS-STKKNVQKEACWTVSNITAGNKDQIQEVLDSGAIPALLHLLETA 314
+ GA+P L+ LL+ S Q+ A ++ N++ +++ + V SGA+P ++H+L+
Sbjct 393 IAASGAIPLLVNLLTISNDSRTQEHAVTSILNLSICQENKGKIVYSSGAVPGIVHVLQKG 452
Query 315 DFDTKKEAAWAVSNAVSGGTDMQVALLVEKGCIPPLCSLL 354
+ ++ AA + + D + G IPPL +LL
Sbjct 453 SMEARENAAATLFSL--SVIDENKVTIGAAGAIPPLVTLL 490
Score = 41.6 bits (96), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 65/270 (24%), Positives = 115/270 (42%), Gaps = 6/270 (2%)
Query 69 AAWALTNIASGTQEQTRVVVECGAIPLFVELLRCPKE-EVREQAVWALGNIAGDSPSCRN 127
AA + +A + GAIPL V LL + +E AV ++ N++ +
Sbjct 375 AAGEIRLLAKQNNHNRVAIAASGAIPLLVNLLTISNDSRTQEHAVTSILNLSICQENKGK 434
Query 128 LVLDGGALPPLLEQLNTPGSKVAMLRNATWTLSNLCRGKPAPVFEKVSPALPALAVLIYS 187
+V GA+P ++ L GS A NA TL +L V + A+P L L+
Sbjct 435 IVYSSGAVPGIVHVLQK-GSMEAR-ENAAATLFSLSVIDENKVTIGAAGAIPPLVTLLSE 492
Query 188 DDAEVLTDACWALSYLSDGPNDRIKAVLEAGVCARLVELLSHQSNLVQTPALRAVGNIVT 247
DA AL L ++ KAV AG+ L+ LL+ + + +L + + +
Sbjct 493 GSQRGKKDAATALFNLCIFQGNKGKAV-RAGLVPVLMRLLTEPESGMVDESLSILAILSS 551
Query 248 GDDTQTQVVLTCGALPKLLTLLSSTKKNVQKEACWTVSNITAGNKDQIQEVLDSGAIPAL 307
D +++V A+P L+ + S ++ + + ++ + N+ + E G + L
Sbjct 552 HPDGKSEVG-AADAVPVLVDFIRSGSPRNKENSAAVLVHLCSWNQQHLIEAQKLGIMDLL 610
Query 308 LHLLETADFDTKKEAAWAVSNAVSGGTDMQ 337
+ + E K++AA + N S D Q
Sbjct 611 IEMAENGTDRGKRKAAQLL-NRFSRFNDQQ 639
Score = 39.3 bits (90), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 42/178 (23%), Positives = 85/178 (47%), Gaps = 11/178 (6%)
Query 196 ACWALSYLSDGPNDRIKAVLEAGVCARLVELLS-HQSNLVQTPALRAVGNIVTGDDTQTQ 254
A + L+ N A+ +G LV LL+ + Q A+ ++ N+ + + +
Sbjct 375 AAGEIRLLAKQNNHNRVAIAASGAIPLLVNLLTISNDSRTQEHAVTSILNLSICQENKGK 434
Query 255 VVLTCGALPKLLTLLSSTKKNVQKEACWTVSNITAGNKDQIQEVLDSGAIPALLHLLETA 314
+V + GA+P ++ +L ++ A T+ +++ +++++ + +GAIP L+ LL
Sbjct 435 IVYSSGAVPGIVHVLQKGSMEARENAAATLFSLSVIDENKVT-IGAAGAIPPLVTLLSEG 493
Query 315 DFDTKKEAAWAVSN-AVSGGTDMQVALLVEKGCIPPLCSLL-----GCMDNKIIMVAL 366
KK+AA A+ N + G + V G +P L LL G +D + ++A+
Sbjct 494 SQRGKKDAATALFNLCIFQGNKGKA---VRAGLVPVLMRLLTEPESGMVDESLSILAI 548
> At3g01400
Length=355
Score = 47.8 bits (112), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 46/178 (25%), Positives = 87/178 (48%), Gaps = 8/178 (4%)
Query 186 YSDDAEVLTDACWALSYLS-DGPNDRIKAVLEAGVCARLVELLSHQSNLVQTPALRAVGN 244
YS D + A + LS + P +RIK + +AG L+ L+S +Q + A+ N
Sbjct 75 YSIDEQ--KQAAMEIRLLSKNKPENRIK-IAKAGAIKPLISLISSSDLQLQEYGVTAILN 131
Query 245 IVTGDDTQTQVVLTCGALPKLLTLLSSTKKNVQKEACWTVSNITAGNKDQIQEVLDSGAI 304
+ D+ + + + GA+ L+ L ++ A + ++ ++++ + SGAI
Sbjct 132 LSLCDENKESIA-SSGAIKPLVRALKMGTPTAKENAACALLRLSQIEENKVA-IGRSGAI 189
Query 305 PALLHLLETADFDTKKEAAWAVSNAVSGGTDMQVALLVEKGCIPPLCSLLGCMDNKII 362
P L++LLET F KK+A+ A+ + S + A V+ G + PL L+ + ++
Sbjct 190 PLLVNLLETGGFRAKKDASTALYSLCSAKENKIRA--VQSGIMKPLVELMADFGSNMV 245
Score = 44.3 bits (103), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 63/276 (22%), Positives = 119/276 (43%), Gaps = 11/276 (3%)
Query 50 VPHFVRFLSMHGRYQLQFEAAWALTNIASGTQEQTRVVVECGAIPLFVELLRCPKEEVRE 109
+ H V L Q +AA + ++ E + + GAI + L+ +++E
Sbjct 64 INHLVSHLDSSYSIDEQKQAAMEIRLLSKNKPENRIKIAKAGAIKPLISLISSSDLQLQE 123
Query 110 QAVWALGNIAGDSPSCRNLVLDGGALPPLLE--QLNTPGSKVAMLRNATWTLSNLCRGKP 167
V A+ N++ + + + GA+ PL+ ++ TP +K NA L L + +
Sbjct 124 YGVTAILNLSLCDEN-KESIASSGAIKPLVRALKMGTPTAK----ENAACALLRLSQIEE 178
Query 168 APVFEKVSPALPALAVLIYSDDAEVLTDACWALSYLSDGPNDRIKAVLEAGVCARLVELL 227
V S A+P L L+ + DA AL L ++I+AV ++G+ LVEL+
Sbjct 179 NKVAIGRSGAIPLLVNLLETGGFRAKKDASTALYSLCSAKENKIRAV-QSGIMKPLVELM 237
Query 228 S-HQSNLVQTPALRAVGNIVTGDDTQTQVVLTCGALPKLLTLLSSTKKNVQKEACWTVSN 286
+ SN+V A V +++ ++ G +P L+ ++ + ++ A +
Sbjct 238 ADFGSNMVDKSAF--VMSLLMSVPESKPAIVEEGGVPVLVEIVEVGTQRQKEMAVSILLQ 295
Query 287 ITAGNKDQIQEVLDSGAIPALLHLLETADFDTKKEA 322
+ + V GAIP L+ L + K++A
Sbjct 296 LCEESVVYRTMVAREGAIPPLVALSQAGTSRAKQKA 331
Score = 38.9 bits (89), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 44/184 (23%), Positives = 72/184 (39%), Gaps = 45/184 (24%)
Query 7 LQQLVQALASGVPDAELEGTQLFRRALSLENNPPIQEVIDAGAVPHFVRFLSMHGRYQLQ 66
++ LV+AL G P A+ R +E N + +GA+P V L G ++ +
Sbjct 148 IKPLVRALKMGTPTAKENAACALLRLSQIEENKVA--IGRSGAIPLLVNLLETGG-FRAK 204
Query 67 FEAAWALTNIASGTQEQTRVV--------------------------------------- 87
+A+ AL ++ S + + R V
Sbjct 205 KDASTALYSLCSAKENKIRAVQSGIMKPLVELMADFGSNMVDKSAFVMSLLMSVPESKPA 264
Query 88 -VECGAIPLFVELLRCPKEEVREQAVWALGNIAGDSPSCRNLVLDGGALPPL--LEQLNT 144
VE G +P+ VE++ + +E AV L + +S R +V GA+PPL L Q T
Sbjct 265 IVEEGGVPVLVEIVEVGTQRQKEMAVSILLQLCEESVVYRTMVAREGAIPPLVALSQAGT 324
Query 145 PGSK 148
+K
Sbjct 325 SRAK 328
> SPBC354.14c
Length=550
Score = 47.8 bits (112), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 80/308 (25%), Positives = 138/308 (44%), Gaps = 12/308 (3%)
Query 56 FLSMHGRYQLQFEAAWALTNIASGTQEQTRVVVECGAIPLFVELLRCPKEEVREQAVWAL 115
L+ LQ AA A I T++ R V P+ LL+ P E++ A AL
Sbjct 54 ILAYSDNLDLQRSAALAFAEI---TEKDVREVDRETIEPVLF-LLQSPDAEIQRAASVAL 109
Query 116 GNIAGDSPSCRNLVLDGGALPPLLEQLNTPGSKVAMLRNATWTLSNLCRGKPAPVFEKVS 175
GN+A ++ + + LV+ L L+ Q+ +P +V NA ++NL S
Sbjct 110 GNLAVNAEN-KALVVKLNGLDLLIRQMMSPHVEVQC--NAVGCITNLATLDENKSKIAHS 166
Query 176 PALPALAVLIYSDDAEVLTDACWALSYLSDGPNDRIKAVLEAGVCARLVELLSHQSNLVQ 235
AL L L S D V +A AL ++ +R + ++ AG LV LL VQ
Sbjct 167 GALGPLTRLAKSKDIRVQRNATGALLNMTHSYENR-QQLVSAGTIPVLVSLLPSSDTDVQ 225
Query 236 TPALRAVGNIVTGDDTQTQVVLTCGALPK-LLTLLSSTKKNVQKEACWTVSNITAGNKDQ 294
++ NI + ++ + L + L+ L+ ++ VQ +A + N+ + + Q
Sbjct 226 YYCTTSISNIAVDAVHRKRLAQSEPKLVRSLIQLMDTSSPKVQCQAALALRNLASDERYQ 285
Query 295 IQEVLDSGAIPALLHLLETADFDTKKEAAWAVSNAVSGGTDMQVALLVEKGCIPPLCSLL 354
I E++ S A+P+LL LL ++ + + N + + +++ G + PL LL
Sbjct 286 I-EIVQSNALPSLLRLLRSSYLPLILASVACIRNI--SIHPLNESPIIDAGFLRPLVDLL 342
Query 355 GCMDNKII 362
C +N+ I
Sbjct 343 SCTENEEI 350
Score = 40.4 bits (93), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 80/338 (23%), Positives = 147/338 (43%), Gaps = 16/338 (4%)
Query 2 LEGVDLQQLVQALASGVPDAELEGTQLFRRALSLENNPPIQEVIDAGAVPHFVRFLSMHG 61
L G+DL L++ + S + + +L+ N ++ +GA+ R L+
Sbjct 125 LNGLDL--LIRQMMSPHVEVQCNAVGCITNLATLDENK--SKIAHSGALGPLTR-LAKSK 179
Query 62 RYQLQFEAAWALTNIASGTQEQTRVVVECGAIPLFVELLRCPKEEVREQAVWALGNIAGD 121
++Q A AL N+ + + E + +V G IP+ V LL +V+ ++ NIA D
Sbjct 180 DIRVQRNATGALLNM-THSYENRQQLVSAGTIPVLVSLLPSSDTDVQYYCTTSISNIAVD 238
Query 122 SPSCRNLVLDGGAL-PPLLEQLNTPGSKVAMLRNATWTLSNLCRGKPAPVFEKVSPALPA 180
+ + L L L++ ++T KV A L NL + + S ALP+
Sbjct 239 AVHRKRLAQSEPKLVRSLIQLMDTSSPKVQC--QAALALRNLASDERYQIEIVQSNALPS 296
Query 181 LAVLIYSDDAEVLTDACWALSYLSDGPNDRIKAVLEAGVCARLVELLSHQSN-LVQTPAL 239
L L+ S ++ + + +S P + +++AG LV+LLS N +Q A+
Sbjct 297 LLRLLRSSYLPLILASVACIRNISIHPLNE-SPIIDAGFLRPLVDLLSCTENEEIQCHAV 355
Query 240 RAVGNIVTGDDTQTQVVLTCGALPKLLTLLSSTKKNVQKE--ACWTVSNITAGNKDQIQE 297
+ N+ + + ++ A+ KL L+ +VQ E AC V ++ K +
Sbjct 356 STLRNLAASSERNKRAIIEANAIQKLRCLILDAPVSVQSEMTACLAVLALSDEFKSYL-- 413
Query 298 VLDSGAIPALLHLLETADFDTKKEAAWAVSNAVSGGTD 335
L+ G L+ L ++ + + +A A+ N S D
Sbjct 414 -LNFGICNVLIPLTDSMSIEVQGNSAAALGNLSSNVDD 450
Score = 40.4 bits (93), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 78/328 (23%), Positives = 133/328 (40%), Gaps = 76/328 (23%)
Query 42 QEVIDAGAVPHFVRFLSMHGRYQLQFEAAWALTNIASGTQEQTRVV-VECGAIPLFVELL 100
Q+++ AG +P V L +Q+ +++NIA + R+ E + ++L+
Sbjct 202 QQLVSAGTIPVLVSLLP-SSDTDVQYYCTTSISNIAVDAVHRKRLAQSEPKLVRSLIQLM 260
Query 101 RCPKEEVREQAVWALGNIAGD------------------------------SPSC-RNL- 128
+V+ QA AL N+A D S +C RN+
Sbjct 261 DTSSPKVQCQAALALRNLASDERYQIEIVQSNALPSLLRLLRSSYLPLILASVACIRNIS 320
Query 129 --------VLDGGALPPLLEQLN-TPGSKVAMLRNATWTLSNLC----RGKPAPVFEKVS 175
++D G L PL++ L+ T ++ +A TL NL R K A + +
Sbjct 321 IHPLNESPIIDAGFLRPLVDLLSCTENEEIQC--HAVSTLRNLAASSERNKRAII---EA 375
Query 176 PALPALAVLIYSDDAEVLTD--ACWALSYLSDGPNDRIKAVLEAGVCARLVELLSHQSNL 233
A+ L LI V ++ AC A+ LSD + +L G+C L+ L S
Sbjct 376 NAIQKLRCLILDAPVSVQSEMTACLAVLALSD---EFKSYLLNFGICNVLIPLTDSMSIE 432
Query 234 VQTPALRAVGNIVTGDDTQTQVVLTC------GALPKLLTLLSSTKKNVQKEACWTVSNI 287
VQ + A+GN+ + D ++ + C G L+ LSS A WT
Sbjct 433 VQGNSAAALGNLSSNVDDYSRFI-ECWDSPAGGIHGYLVRFLSSEDSTFAHIAAWT---- 487
Query 288 TAGNKDQIQEVLDSGAIPALLHLLETAD 315
I ++L++G +P L ++++D
Sbjct 488 -------IVQLLEAG-VPRLTAFIQSSD 507
> At5g67340
Length=698
Score = 47.4 bits (111), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 38/114 (33%), Positives = 60/114 (52%), Gaps = 7/114 (6%)
Query 245 IVTGDDTQTQVVLT-CGALPKLLTLLSSTKKNVQKEA--CWTVSNITAGNKDQIQEVLDS 301
I+ + T ++V+ C A+P L++LL ST + +Q +A C +I NK I E S
Sbjct 443 ILARNSTDNRIVIARCEAIPSLVSLLYSTDERIQADAVTCLLNLSINDNNKSLIAE---S 499
Query 302 GAIPALLHLLETADFDTKKEAAWAVSNAVSGGTDMQVALLVEKGCIPPLCSLLG 355
GAI L+H+L+T + K + A ++S + + + E G I PL LLG
Sbjct 500 GAIVPLIHVLKTGYLEEAKANSAATLFSLSVIEEYKTE-IGEAGAIEPLVDLLG 552
Score = 45.8 bits (107), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 40/124 (32%), Positives = 60/124 (48%), Gaps = 9/124 (7%)
Query 16 SGVPDAELEGTQLFRRALSLENNPPIQEVIDAGAVPHFVRFLSMHGRYQLQFEAAWALTN 75
SG DA T LF ++ EN +VI+AGAV + V M + + +A L N
Sbjct 557 SGKKDA---ATALFNLSIHHENK---TKVIEAGAVRYLVEL--MDPAFGMVEKAVVVLAN 608
Query 76 IASGTQEQTRVVVECGAIPLFVELLRCPKEEVREQAVWALGNIAGDSPSCRNLVLDGGAL 135
+A+ +E + E G IP+ VE++ +E A AL + SP N V+ G +
Sbjct 609 LAT-VREGKIAIGEEGGIPVLVEVVELGSARGKENATAALLQLCTHSPKFCNNVIREGVI 667
Query 136 PPLL 139
PPL+
Sbjct 668 PPLV 671
Score = 45.4 bits (106), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 62/248 (25%), Positives = 110/248 (44%), Gaps = 5/248 (2%)
Query 66 QFEAAWALTNIASGTQEQTRVVVECGAIPLFVELLRCPKEEVREQAVWALGNIAGDSPSC 125
Q EA + +A + + V+ C AIP V LL E ++ AV L N++ + +
Sbjct 434 QREATARIRILARNSTDNRIVIARCEAIPSLVSLLYSTDERIQADAVTCLLNLSINDNN- 492
Query 126 RNLVLDGGALPPLLEQLNTPGSKVAMLRNATWTLSNLCRGKPAPVFEKVSPALPALAVLI 185
++L+ + GA+ PL+ L T G N+ TL +L + + A+ L L+
Sbjct 493 KSLIAESGAIVPLIHVLKT-GYLEEAKANSAATLFSLSVIEEYKTEIGEAGAIEPLVDLL 551
Query 186 YSDDAEVLTDACWALSYLSDGPNDRIKAVLEAGVCARLVELLSHQSNLVQTPALRAVGNI 245
S DA AL LS ++ K V+EAG LVEL+ +V+ A+ + N+
Sbjct 552 GSGSLSGKKDAATALFNLSIHHENKTK-VIEAGAVRYLVELMDPAFGMVEK-AVVVLANL 609
Query 246 VTGDDTQTQVVLTCGALPKLLTLLSSTKKNVQKEACWTVSNITAGNKDQIQEVLDSGAIP 305
T + + + G +P L+ ++ ++ A + + + V+ G IP
Sbjct 610 ATVREGKIAIGEE-GGIPVLVEVVELGSARGKENATAALLQLCTHSPKFCNNVIREGVIP 668
Query 306 ALLHLLET 313
L+ L ++
Sbjct 669 PLVALTKS 676
Score = 37.4 bits (85), Expect = 0.054, Method: Compositional matrix adjust.
Identities = 42/181 (23%), Positives = 97/181 (53%), Gaps = 11/181 (6%)
Query 177 ALPALAVLIYSDDAEVLTDACWALSYLSDGPNDRIKAVLEAGVCARLVELLSHQSNLVQT 236
A+P+L L+YS D + DA L LS N++ + E+G L+ +L ++ ++
Sbjct 460 AIPSLVSLLYSTDERIQADAVTCLLNLSINDNNK-SLIAESGAIVPLIHVL--KTGYLEE 516
Query 237 PALRAVGNIVT---GDDTQTQVVLTCGALPKLLTLLSSTKKNVQKEACWTVSNITAGNKD 293
+ + + ++ +T++ GA+ L+ LL S + +K+A + N++ +++
Sbjct 517 AKANSAATLFSLSVIEEYKTEIG-EAGAIEPLVDLLGSGSLSGKKDAATALFNLSIHHEN 575
Query 294 QIQEVLDSGAIPALLHLLETADFDTKKEAAWAVSNAVSGGTDMQVALLVEKGCIPPLCSL 353
+ + V+++GA+ L+ L++ A F ++A ++N ++ + ++A + E+G IP L +
Sbjct 576 KTK-VIEAGAVRYLVELMDPA-FGMVEKAVVVLAN-LATVREGKIA-IGEEGGIPVLVEV 631
Query 354 L 354
+
Sbjct 632 V 632
> At3g20170
Length=475
Score = 47.0 bits (110), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 60/263 (22%), Positives = 110/263 (41%), Gaps = 18/263 (6%)
Query 71 WALTNIASG---TQEQTRVVVECGAIPLFVELLRCPKEEVREQAVWALGNIAGDSPSCRN 127
W L I S +E RV+V G + VE + RE+A A+G I G + R
Sbjct 170 WYLLEILSALTTIRESRRVLVHSGGLKFLVEAAKVGNLASRERACHAIGLI-GVTRRARR 228
Query 128 LVLDGGALPPLLEQLNTPGSKVAMLRNATWTLSNLCRGKPAPVFEKVSPALPALAVLIYS 187
++++ G +P L++ K +L + + PV E + ++P L+
Sbjct 229 ILVEAGVIPALVDLYRDGDDKAKLLAGNALGIISAQTEYIRPVTE--AGSIPLYVELLSG 286
Query 188 DD--AEVLTDACWALSYLSDGPNDRIKAVLEAGVCARLVELLSHQSNLVQTPALRAVGNI 245
D + + + + + +++G AVL + +LV +L N + A + ++
Sbjct 287 QDPMGKDIAEDVFCILAVAEG-----NAVL---IAEQLVRILRAGDNEAKLAASDVLWDL 338
Query 246 VTGDDTQTQVVLTCGALPKLLTLLSSTKKNVQKEACWTVSNITAGNKDQIQEVLDSGAIP 305
G V+ GA+P L+ LL ++ +S ++ D+ + DSG IP
Sbjct 339 -AGYRHSVSVIRGSGAIPLLIELLRDGSLEFRERISGAISQLSYNENDR-EAFSDSGMIP 396
Query 306 ALLHLLETADFDTKKEAAWAVSN 328
L+ L + + AA A+ N
Sbjct 397 ILIEWLGDESEELRDNAAEALIN 419
Score = 38.5 bits (88), Expect = 0.025, Method: Compositional matrix adjust.
Identities = 58/214 (27%), Positives = 91/214 (42%), Gaps = 15/214 (7%)
Query 44 VIDAGAVPHFVRFLSMHGRYQLQFEAAWALTNIASGTQEQTRVVVECGAIPLFVELLRCP 103
+++AG +P V L G + + A AL I S E R V E G+IPL+VELL
Sbjct 230 LVEAGVIPALVD-LYRDGDDKAKLLAGNAL-GIISAQTEYIRPVTEAGSIPLYVELLSGQ 287
Query 104 KEEVREQAVWALGNIAGDSPSCRNLVLDGGA--LPPLLEQLNTPGSKVAMLRNATWTLSN 161
+G + C V +G A + L ++ G A L A+ L +
Sbjct 288 D---------PMGKDIAEDVFCILAVAEGNAVLIAEQLVRILRAGDNEAKL-AASDVLWD 337
Query 162 LCRGKPAPVFEKVSPALPALAVLIYSDDAEVLTDACWALSYLSDGPNDRIKAVLEAGVCA 221
L + + + S A+P L L+ E A+S LS NDR +A ++G+
Sbjct 338 LAGYRHSVSVIRGSGAIPLLIELLRDGSLEFRERISGAISQLSYNENDR-EAFSDSGMIP 396
Query 222 RLVELLSHQSNLVQTPALRAVGNIVTGDDTQTQV 255
L+E L +S ++ A A+ N + +V
Sbjct 397 ILIEWLGDESEELRDNAAEALINFSEDQEHYARV 430
Score = 32.7 bits (73), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 29/120 (24%), Positives = 57/120 (47%), Gaps = 16/120 (13%)
Query 8 QQLVQALASGVPDAELEGTQL------FRRALSLENNPPIQEVIDAGAVPHFVRFLSMHG 61
+QLV+ L +G +A+L + + +R ++S+ + +GA+P + L G
Sbjct 314 EQLVRILRAGDNEAKLAASDVLWDLAGYRHSVSV--------IRGSGAIPLLIELLR-DG 364
Query 62 RYQLQFEAAWALTNIASGTQEQTRVVVECGAIPLFVELLRCPKEEVREQAVWALGNIAGD 121
+ + + A++ + S + + G IP+ +E L EE+R+ A AL N + D
Sbjct 365 SLEFRERISGAISQL-SYNENDREAFSDSGMIPILIEWLGDESEELRDNAAEALINFSED 423
Score = 30.4 bits (67), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 28/108 (25%), Positives = 50/108 (46%), Gaps = 3/108 (2%)
Query 292 KDQIQEVLDSGAIPALLHLLETADFDTKKEAAWAVSNAVSGGTDMQVALLVEKGCIPPLC 351
++ + ++ SG + L+ + + +++ A A+ + G T +LVE G IP L
Sbjct 183 RESRRVLVHSGGLKFLVEAAKVGNLASRERACHAI--GLIGVTRRARRILVEAGVIPALV 240
Query 352 SLLGCMDNKIIMVALEALNAILRTGKKIKE-TEGLSVNPYATLIEQAD 398
L D+K ++A AL I + I+ TE S+ Y L+ D
Sbjct 241 DLYRDGDDKAKLLAGNALGIISAQTEYIRPVTEAGSIPLYVELLSGQD 288
> Hs22054575
Length=191
Score = 46.6 bits (109), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 38/168 (22%), Positives = 81/168 (48%), Gaps = 6/168 (3%)
Query 167 PAPVFEKV---SPALPALAVLIYSDDAEVLTDACWALSYLSDGPNDRIKAVLEAGVCARL 223
P VF+ + S + +++ S + E+L AC A+ + + +LE G L
Sbjct 13 PKDVFDPLMIESKKAATVVLMLNSPEEEILAKACEAIYKFALKGEENKTTLLELGAVEPL 72
Query 224 VELLSHQSNLVQTPALRAVGNIVTGDDTQTQVVLTCGALPKLLTLLSSTKKNVQKE-ACW 282
+LL+H+ +V+ A G + + +D + +++ + ++ L+ ++ V E A
Sbjct 73 TKLLTHEDKIVRRNATMIFGILASNNDVK-KLLRELDVMNSVIAQLAPEEEVVIHEFASL 131
Query 283 TVSNITAGNKDQIQEVLDSGAIPALLHLLETADFDTKKEAAWAVSNAV 330
++N++A ++Q + + G + L+ LL + D D KK + + N V
Sbjct 132 CLANMSAEYTSKVQ-IFEHGGLEPLIRLLSSPDPDVKKNSMECIYNLV 178
> At4g38610
Length=1083
Score = 46.2 bits (108), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 45/157 (28%), Positives = 73/157 (46%), Gaps = 6/157 (3%)
Query 50 VPHFVRFLSMHGRYQLQFEAAWALTNIASGTQEQTRVVVECGAIPLFV-ELLRCPKEEVR 108
VP V L+ + AA ALT++ VV GA+ V LL ++
Sbjct 239 VPVLVGLLNHESNPDIMLLAARALTHLCDVLPSSCAAVVHYGAVSCLVARLLTIEYMDLA 298
Query 109 EQAVWALGNIAGDSPSCRNLVLDGGALPPLLEQLNTPGSKVAMLRNATWTLSNLCRGKPA 168
EQ++ AL I+ + P+ L GAL +L L+ + V R A T +N+C+ P+
Sbjct 299 EQSLQALKKISQEHPTA---CLRAGALMAVLSYLDFFSTGVQ--RVALSTAANMCKKLPS 353
Query 169 PVFEKVSPALPALAVLIYSDDAEVLTDACWALSYLSD 205
+ V A+P L L+ D++VL A L+ +++
Sbjct 354 DASDYVMEAVPLLTNLLQYHDSKVLEYASICLTRIAE 390
Lambda K H
0.316 0.132 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 10664741630
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40