bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0548_orf3
Length=165
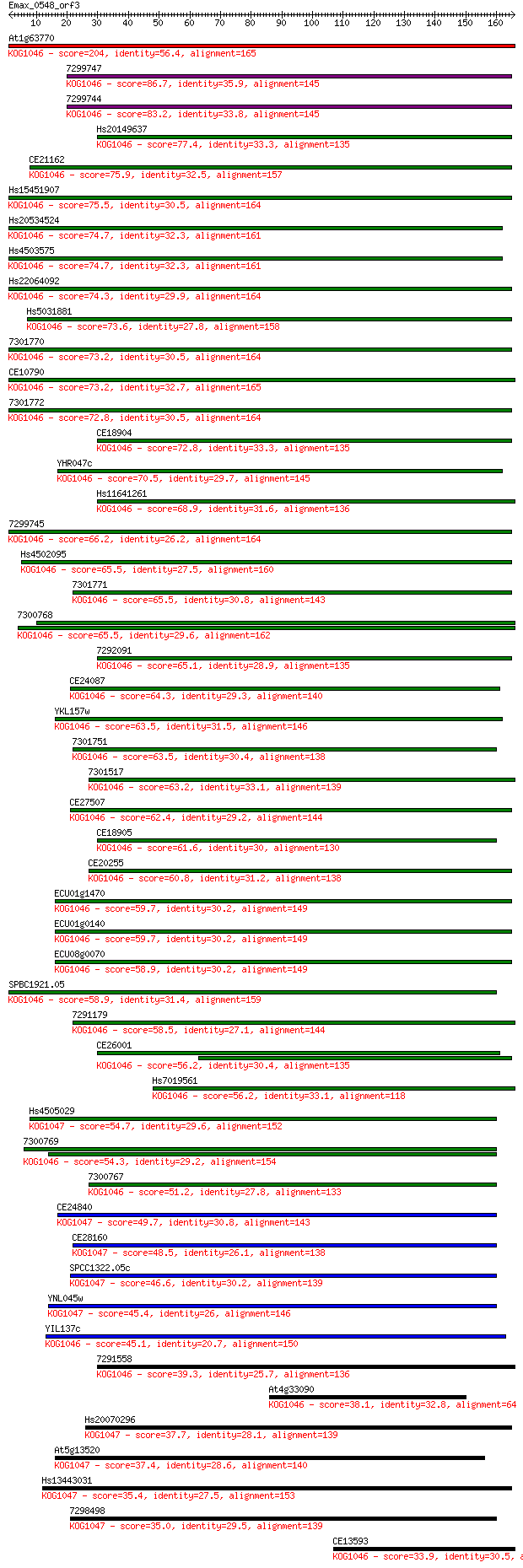
Score E
Sequences producing significant alignments: (Bits) Value
At1g63770 204 8e-53
7299747 86.7 2e-17
7299744 83.2 2e-16
Hs20149637 77.4 1e-14
CE21162 75.9 4e-14
Hs15451907 75.5 5e-14
Hs20534524 74.7 7e-14
Hs4503575 74.7 7e-14
Hs22064092 74.3 1e-13
Hs5031881 73.6 2e-13
7301770 73.2 2e-13
CE10790 73.2 2e-13
7301772 72.8 3e-13
CE18904 72.8 3e-13
YHR047c 70.5 1e-12
Hs11641261 68.9 4e-12
7299745 66.2 3e-11
Hs4502095 65.5 5e-11
7301771 65.5 5e-11
7300768 65.5 5e-11
7292091 65.1 5e-11
CE24087 64.3 1e-10
YKL157w 63.5 2e-10
7301751 63.5 2e-10
7301517 63.2 2e-10
CE27507 62.4 4e-10
CE18905 61.6 7e-10
CE20255 60.8 1e-09
ECU01g1470 59.7 3e-09
ECU01g0140 59.7 3e-09
ECU08g0070 58.9 4e-09
SPBC1921.05 58.9 4e-09
7291179 58.5 5e-09
CE26001 56.2 3e-08
Hs7019561 56.2 3e-08
Hs4505029 54.7 8e-08
7300769 54.3 1e-07
7300767 51.2 8e-07
CE24840 49.7 3e-06
CE28160 48.5 5e-06
SPCC1322.05c 46.6 2e-05
YNL045w 45.4 5e-05
YIL137c 45.1 7e-05
7291558 39.3 0.004
At4g33090 38.1 0.008
Hs20070296 37.7 0.010
At5g13520 37.4 0.013
Hs13443031 35.4 0.055
7298498 35.0 0.072
CE13593 33.9 0.14
> At1g63770
Length=964
Score = 204 bits (518), Expect = 8e-53, Method: Compositional matrix adjust.
Identities = 93/165 (56%), Positives = 122/165 (73%), Gaps = 2/165 (1%)
Query 1 SNGNKTEEGPVQDSPDRHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVY 60
SNGN +G ++ RH+ ++ DP KP YLFA++AG + DTFTT+SG++V+L ++
Sbjct 238 SNGNLISQGDIEGG--RHYALWEDPFKKPCYLFALVAGQLVSRDDTFTTRSGRQVSLKIW 295
Query 61 SEPEQMHKLNWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCS 120
+ E + K AM+S+K +MKW+ED FG EYDLD FN+ V DFN GAMENK LNIFN
Sbjct 296 TPAEDLPKTAHAMYSLKAAMKWDEDVFGLEYDLDLFNIVAVPDFNMGAMENKSLNIFNSK 355
Query 121 LLLASMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTLK 165
L+LAS +T+TD D+ +L V+GHEYFHNWTGNRVT RDWFQL+LK
Sbjct 356 LVLASPETATDADYAAILGVIGHEYFHNWTGNRVTCRDWFQLSLK 400
> 7299747
Length=985
Score = 86.7 bits (213), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 52/145 (35%), Positives = 73/145 (50%), Gaps = 3/145 (2%)
Query 20 VVFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWAMHSVKVS 79
V F + P +YL A + DF TT G +AL VY+ P Q+ K +A+ +
Sbjct 328 VTFAETVPMSTYLAAFVVSDFQY---KETTVEGTSIALKVYAPPAQVEKTQYALDTAAGV 384
Query 80 MKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTSTDDDFERVLA 139
M + ++F Y L ++ + DF +GAMEN GL F + LL TS+ + +RV
Sbjct 385 MAYYINYFNVSYALPKLDLVAIPDFVSGAMENWGLVTFRETALLYDESTSSSVNKQRVAI 444
Query 140 VVGHEYFHNWTGNRVTVRDWFQLTL 164
VV HE H W GN VT+ W L L
Sbjct 445 VVAHELAHQWFGNLVTMNWWNDLWL 469
> 7299744
Length=988
Score = 83.2 bits (204), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 49/146 (33%), Positives = 80/146 (54%), Gaps = 1/146 (0%)
Query 20 VVFTDPHPKPSYLFAILAGDFAAIVDTFTTKS-GKEVALAVYSEPEQMHKLNWAMHSVKV 78
V F P +YL + DFA + TK G+ +++VY+ PEQ+ K++ A+ K
Sbjct 302 VTFAKSVPMSTYLACFIVSDFAYKQVSIDTKGIGETFSMSVYATPEQLDKVDLAVTIGKG 361
Query 79 SMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTSTDDDFERVL 138
+++ D+F Y L ++A + DF +GAME+ GL + + LL TS+ + +R+
Sbjct 362 VIEYYIDYFQIAYPLPKLDMAAIPDFVSGAMEHWGLVTYRETSLLYDEATSSATNKQRIA 421
Query 139 AVVGHEYFHNWTGNRVTVRDWFQLTL 164
+V+ HE+ H W GN VT+ W L L
Sbjct 422 SVIAHEFAHMWFGNLVTMNWWNDLWL 447
> Hs20149637
Length=948
Score = 77.4 bits (189), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 45/135 (33%), Positives = 75/135 (55%), Gaps = 4/135 (2%)
Query 30 SYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWAMHSVKVSMKWEEDHFGR 89
+YL A + DF ++ + TKSG V ++VY+ P+++++ ++A+ + +++ ED+F
Sbjct 244 TYLVAFIISDFESV--SKITKSG--VKVSVYAVPDKINQADYALDAAVTLLEFYEDYFSI 299
Query 90 EYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTSTDDDFERVLAVVGHEYFHNW 149
Y L ++A + DF +GAMEN GL + S LL + S+ + V HE H W
Sbjct 300 PYPLPKQDLAAIPDFQSGAMENWGLTTYRESALLFDAEKSSASSKLGITVTVAHELAHQW 359
Query 150 TGNRVTVRDWFQLTL 164
GN VT+ W L L
Sbjct 360 FGNLVTMEWWNDLWL 374
> CE21162
Length=988
Score = 75.9 bits (185), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 51/158 (32%), Positives = 78/158 (49%), Gaps = 5/158 (3%)
Query 8 EGPVQDSPDRHFVVFTDPHPK-PSYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQM 66
E V+D + P P+ SYL AI +F + TTKSG V V+S PE+
Sbjct 264 EDKVEDGQPGFIISTFKPTPRMSSYLLAIFISEFE--YNEATTKSG--VRFRVWSRPEEK 319
Query 67 HKLNWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASM 126
+ +A+ + +++ E ++ + L ++ + DF+AGAMEN GL + S LL
Sbjct 320 NSTMYAVEAGVKCLEYYEKYYNISFPLPKQDMVALPDFSAGAMENWGLITYRESALLYDP 379
Query 127 QTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTL 164
+ + RV V+ HE H W GN VT++ W L L
Sbjct 380 RIYSGSQKRRVAVVIAHELAHQWFGNLVTLKWWNDLWL 417
> Hs15451907
Length=875
Score = 75.5 bits (184), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 50/164 (30%), Positives = 77/164 (46%), Gaps = 4/164 (2%)
Query 1 SNGNKTEEGPVQDSPDRHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVY 60
SN N + P D + V F +YL A + G++ D T+S V + VY
Sbjct 170 SNMNVIDRKPYPDDENLVEVKFARTPVMSTYLVAFVVGEY----DFVETRSKDGVCVRVY 225
Query 61 SEPEQMHKLNWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCS 120
+ + + +A+ ++ + +D+F Y L ++ ++DF AGAMEN GL + +
Sbjct 226 TPVGKAEQGKFALEVAAKTLPFYKDYFNVPYPLPKIDLIAIADFAAGAMENWGLVTYRET 285
Query 121 LLLASMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTL 164
LL + S + V VVGHE H W GN VT+ W L L
Sbjct 286 ALLIDPKNSCSSSRQWVALVVGHELAHQWFGNLVTMEWWTHLWL 329
> Hs20534524
Length=957
Score = 74.7 bits (182), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 52/161 (32%), Positives = 74/161 (45%), Gaps = 7/161 (4%)
Query 1 SNGNKTEEGPVQDSPDRHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVY 60
SN +E V D R F P +YL F ++ + SGK L +Y
Sbjct 257 SNMPVAKEESVDDKWTR--TTFEKSVPMSTYLVCFAVHQFDSV--KRISNSGK--PLTIY 310
Query 61 SEPEQMHKLNWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCS 120
+PEQ H +A + K + E++F Y L + + DF GAMEN GL + +
Sbjct 311 VQPEQKHTAEYAANITKSVFDYFEEYFAMNYSLPKLDKIAIPDFGTGAMENWGLITYRET 370
Query 121 LLLASMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQ 161
LL + S + +RV VV HE H W GN VT+ DW++
Sbjct 371 NLLYDPKESASSNQQRVATVVAHELVHQWFGNIVTM-DWWE 410
> Hs4503575
Length=957
Score = 74.7 bits (182), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 52/161 (32%), Positives = 74/161 (45%), Gaps = 7/161 (4%)
Query 1 SNGNKTEEGPVQDSPDRHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVY 60
SN +E V D R F P +YL F ++ + SGK L +Y
Sbjct 257 SNMPVAKEESVDDKWTR--TTFEKSVPMSTYLVCFAVHQFDSV--KRISNSGK--PLTIY 310
Query 61 SEPEQMHKLNWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCS 120
+PEQ H +A + K + E++F Y L + + DF GAMEN GL + +
Sbjct 311 VQPEQKHTAEYAANITKSVFDYFEEYFAMNYSLPKLDKIAIPDFGTGAMENWGLITYRET 370
Query 121 LLLASMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQ 161
LL + S + +RV VV HE H W GN VT+ DW++
Sbjct 371 NLLYDPKESASSNQQRVATVVAHELVHQWFGNIVTM-DWWE 410
> Hs22064092
Length=478
Score = 74.3 bits (181), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 49/164 (29%), Positives = 76/164 (46%), Gaps = 4/164 (2%)
Query 1 SNGNKTEEGPVQDSPDRHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVY 60
SN N + P D + V F +YL A + G++ D T+S V + VY
Sbjct 214 SNMNVIDRKPYPDDENLVEVKFARTPVTSTYLVAFVVGEY----DFVETRSKDGVCVCVY 269
Query 61 SEPEQMHKLNWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCS 120
+ + + +A+ ++ + +D+F Y L ++ ++DF AGAMEN L + +
Sbjct 270 TPVGKAEQGKFALEVAAKTLPFYKDYFNVPYPLPKIDLIAIADFAAGAMENWDLVTYRET 329
Query 121 LLLASMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTL 164
LL + S + V VVGHE H W GN VT+ W L L
Sbjct 330 ALLIDPKNSCSSSRQWVALVVGHELAHQWFGNLVTMEWWTHLRL 373
> Hs5031881
Length=1025
Score = 73.6 bits (179), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 44/158 (27%), Positives = 81/158 (51%), Gaps = 12/158 (7%)
Query 7 EEGPVQDSPDRHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQM 66
++G VQD F++ +YL A + G+ + ++ +++Y+ PE +
Sbjct 340 DDGLVQDE-------FSESVKMSTYLVAFIVGEMKNL-----SQDVNGTLVSIYAVPENI 387
Query 67 HKLNWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASM 126
++++A+ + +++ +++F +Y L ++ + DF AGAMEN GL F LL
Sbjct 388 GQVHYALETTVKLLEFFQNYFEIQYPLKKLDLVAIPDFEAGAMENWGLLTFREETLLYDS 447
Query 127 QTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTL 164
TS+ D + V ++ HE H W GN VT++ W L L
Sbjct 448 NTSSMADRKLVTKIIAHELAHQWFGNLVTMKWWNDLWL 485
> 7301770
Length=814
Score = 73.2 bits (178), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 50/164 (30%), Positives = 76/164 (46%), Gaps = 4/164 (2%)
Query 1 SNGNKTEEGPVQDSPDRHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVY 60
SN E P + PD + F + P +YL A DF+ T + +
Sbjct 197 SNMPVNETRPHESIPDYVWTSFEESLPMSTYLVAYSLNDFSHKPSTLPNGT----LFRTW 252
Query 61 SEPEQMHKLNWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCS 120
+ P + + ++A +K+ E+ FG ++ L + V DF+AGAMEN GL F S
Sbjct 253 ARPNAIDQCDYAAELGPKVLKYYEELFGIKFPLPKVDQIAVPDFSAGAMENWGLVTFAES 312
Query 121 LLLASMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTL 164
LL S + S+ + + +V HE H W GN VT++ W L L
Sbjct 313 TLLYSPEYSSQEAKQETANIVAHELAHQWFGNLVTMKWWTDLWL 356
> CE10790
Length=884
Score = 73.2 bits (178), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 54/165 (32%), Positives = 75/165 (45%), Gaps = 5/165 (3%)
Query 1 SNGNKTEEGPVQDSPDRHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVY 60
SN N E P D R V F SYL A G+ I + TKSG V + VY
Sbjct 176 SNMNVISETPTADG-KRKAVTFATSPKMSSYLVAFAVGELEYI--SAQTKSG--VEMRVY 230
Query 61 SEPEQMHKLNWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCS 120
+ P + + +++ + W + F +Y L ++ + DF+ GAMEN GL +
Sbjct 231 TVPGKKEQGQYSLDLSVKCIDWYNEWFDIKYPLPKCDLIAIPDFSMGAMENWGLVTYREI 290
Query 121 LLLASMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTLK 165
LL ++ RV VV HE H W GN VT++ W L LK
Sbjct 291 ALLVDPGVTSTRQKSRVALVVAHELAHLWFGNLVTMKWWTDLWLK 335
> 7301772
Length=932
Score = 72.8 bits (177), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 50/164 (30%), Positives = 76/164 (46%), Gaps = 4/164 (2%)
Query 1 SNGNKTEEGPVQDSPDRHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVY 60
SN + E P + D + F + P +YL A DF+ T + +
Sbjct 204 SNMPEKETKPHETLADYIWCEFQESVPMSTYLVAYSVNDFSHKPSTLPNSA----LFRTW 259
Query 61 SEPEQMHKLNWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCS 120
+ P + + ++A +++ E FG ++ L + V DF+AGAMEN GL +
Sbjct 260 ARPNAIDQCDYAAQFGPKVLQYYEQFFGIKFPLPKIDQIAVPDFSAGAMENWGLVTYREI 319
Query 121 LLLASMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTL 164
LL S S+ D +RV +VV HE H W GN VT++ W L L
Sbjct 320 ALLYSAAHSSLADKQRVASVVAHELAHQWFGNLVTMKWWTDLWL 363
> CE18904
Length=786
Score = 72.8 bits (177), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 45/135 (33%), Positives = 70/135 (51%), Gaps = 7/135 (5%)
Query 30 SYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWAMHSVKVSMKWEEDHFGR 89
SY+ AI GD TK+G V + VYS+P + ++ A++ ++ ++ E FG
Sbjct 225 SYILAIFVGDVQ--FKEAVTKNG--VRIRVYSDPGHIDSVDHALNVSRIVLEGFEKQFGY 280
Query 90 EYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTSTDDDFERVLAVVGHEYFHNW 149
Y++D ++ V +F GAMEN GL + L+ ++ D + VV HE H W
Sbjct 281 PYEMDKLDLIAVYNFRYGAMENWGLIVHQAYTLIENLMPGNTD---IISEVVAHEIAHQW 337
Query 150 TGNRVTVRDWFQLTL 164
GN VT++ W QL L
Sbjct 338 FGNLVTMKFWDQLWL 352
> YHR047c
Length=856
Score = 70.5 bits (171), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 43/145 (29%), Positives = 70/145 (48%), Gaps = 6/145 (4%)
Query 17 RHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWAMHSV 76
+ + F +YL A + D + + + + VYS P +A +
Sbjct 179 KKYTTFNTTPKMSTYLVAFIVADL-----RYVESNNFRIPVRVYSTPGDEKFGQFAANLA 233
Query 77 KVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTSTDDDFER 136
++++ ED F EY L ++ V +F+AGAMEN GL + LL ++ S+ D +R
Sbjct 234 ARTLRFFEDTFNIEYPLPKMDMVAVHEFSAGAMENWGLVTYRVIDLLLDIENSSLDRIQR 293
Query 137 VLAVVGHEYFHNWTGNRVTVRDWFQ 161
V V+ HE H W GN VT+ DW++
Sbjct 294 VAEVIQHELAHQWFGNLVTM-DWWE 317
> Hs11641261
Length=960
Score = 68.9 bits (167), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 43/136 (31%), Positives = 73/136 (53%), Gaps = 4/136 (2%)
Query 30 SYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWAMHSVKVSMKWEEDHFGR 89
+YL A + DF ++ + T SG V +++Y+ P++ ++ ++A+ + + + E +F
Sbjct 261 TYLVAYIVCDFHSL--SGFTSSG--VKVSIYASPDKRNQTHYALQASLKLLDFYEKYFDI 316
Query 90 EYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTSTDDDFERVLAVVGHEYFHNW 149
Y L ++ + DF GAMEN GL + + LL +TS+ D V V+ HE H W
Sbjct 317 YYPLSKLDLIAIPDFAPGAMENWGLITYRETSLLFDPKTSSASDKLWVTRVIAHELAHQW 376
Query 150 TGNRVTVRDWFQLTLK 165
GN VT+ W + LK
Sbjct 377 FGNLVTMEWWNDIWLK 392
> 7299745
Length=913
Score = 66.2 bits (160), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 43/165 (26%), Positives = 79/165 (47%), Gaps = 4/165 (2%)
Query 1 SNGNKTEEGPVQDSPDRHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTKS-GKEVALAV 59
SN +TE + D + +F +YL I+ DFA+ T G++ ++
Sbjct 225 SNMQQTESNYLGDYTE---AIFETSVSMSTYLVCIIVSDFASQSTTVKANGIGEDFSMQA 281
Query 60 YSEPEQMHKLNWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNC 119
Y+ Q++K+ +A+ + ++ ++ Y L ++A + DF +GAME+ GL +
Sbjct 282 YATSHQINKVEFALEFGQAVTEYYIQYYKVPYPLTKLDMAAIPDFASGAMEHWGLVTYRE 341
Query 120 SLLLASMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTL 164
+ LL S+ + + + + HE H W GN VT++ W L L
Sbjct 342 TALLYDPSYSSTANKQSIAGTLAHEIAHQWFGNLVTMKWWNDLWL 386
> Hs4502095
Length=967
Score = 65.5 bits (158), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 44/162 (27%), Positives = 75/162 (46%), Gaps = 6/162 (3%)
Query 5 KTEEGPVQDSPDRHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPE 64
K P+ + P+ + F +YL A + +F D ++ V + +++ P
Sbjct 252 KGPSTPLPEDPNWNVTEFHTTPKMSTYLLAFIVSEF----DYVEKQASNGVLIRIWARPS 307
Query 65 QMHKL--NWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCSLL 122
+ ++A++ + + H+ Y L + + DFNAGAMEN GL + + L
Sbjct 308 AIAAGHGDYALNVTGPILNFFAGHYDTPYPLPKSDQIGLPDFNAGAMENWGLVTYRENSL 367
Query 123 LASMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTL 164
L +S+ + ERV+ V+ HE H W GN VT+ W L L
Sbjct 368 LFDPLSSSSSNKERVVTVIAHELAHQWFGNLVTIEWWNDLWL 409
> 7301771
Length=1082
Score = 65.5 bits (158), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 44/143 (30%), Positives = 67/143 (46%), Gaps = 4/143 (2%)
Query 22 FTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWAMHSVKVSMK 81
F + P +YL A DF+ T + ++ P + + ++A ++
Sbjct 220 FQESVPMSTYLVAYSVNDFSFKPSTLPNGA----LFRTWARPNAIDQCDYAAEFGPKVLQ 275
Query 82 WEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTSTDDDFERVLAVV 141
+ E+ FG Y L + V DF+AGAMEN GL + S LL S S+ D + + V+
Sbjct 276 YYEEFFGIRYPLPKIDQMAVPDFSAGAMENWGLVKYRESTLLYSPTHSSLADKQDLANVI 335
Query 142 GHEYFHNWTGNRVTVRDWFQLTL 164
HE H W GN VT++ W L L
Sbjct 336 AHELAHQWFGNLVTMKWWTDLWL 358
> 7300768
Length=1878
Score = 65.5 bits (158), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 47/157 (29%), Positives = 72/157 (45%), Gaps = 6/157 (3%)
Query 10 PVQDSPDRHFVVFTDPHPKPS-YLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHK 68
PV + V PK S YL A + DF + TT + V+S + +
Sbjct 229 PVNTAATSSGVTAFQTTPKMSTYLVAFIVSDFES-----TTGELNGIRQRVFSRKGKQDQ 283
Query 69 LNWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQT 128
WA+ S + +FG + L + A + DF+AGAMEN GL + + + Q
Sbjct 284 QEWALWSGLLVESSLASYFGVPFALPKLDQAGIPDFSAGAMENWGLATYREQYMWWNKQN 343
Query 129 STDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTLK 165
ST + + ++GHEY H W G+ V+++ W L LK
Sbjct 344 STINLKTNIANIIGHEYAHMWFGDLVSIKWWTYLWLK 380
Score = 61.6 bits (148), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 46/162 (28%), Positives = 66/162 (40%), Gaps = 5/162 (3%)
Query 4 NKTEEGPVQDSPDRHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEP 63
N PV S VF PSYL A + +F F+ + V+S
Sbjct 1147 NAISNMPVDSSSTSGVTVFQKTVNMPSYLVAFIVSEFV-----FSEGELNGLPQRVFSRN 1201
Query 64 EQMHKLNWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLL 123
H+ WA+ + + K +F + L + A + DF AGAMEN GL + LL
Sbjct 1202 GTEHEQEWALTTGMLVEKRLSGYFDVPFALPKLDQAGIPDFAAGAMENWGLATYREEYLL 1261
Query 124 ASMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTLK 165
+ + ST + + HE H W G+ V + W L LK
Sbjct 1262 YNTENSTTSTQTNIATIEAHEDAHMWFGDLVAIEWWSFLWLK 1303
> 7292091
Length=866
Score = 65.1 bits (157), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 39/135 (28%), Positives = 68/135 (50%), Gaps = 4/135 (2%)
Query 30 SYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWAMHSVKVSMKWEEDHFGR 89
+YL A++ G++ D KS V + V++ + + +A+ + + +D+F
Sbjct 197 TYLVAVVVGEY----DYVEGKSDDGVLVRVFTPVGKREQGTFALEVATKVLPYYKDYFNI 252
Query 90 EYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTSTDDDFERVLAVVGHEYFHNW 149
Y L ++ +SDF+AGAMEN GL + + +L + ++ + + VGHE H W
Sbjct 253 AYPLPKMDLIAISDFSAGAMENWGLVTYRETFVLVDPKNTSLMRKQSIALTVGHEIAHQW 312
Query 150 TGNRVTVRDWFQLTL 164
GN VT+ W L L
Sbjct 313 FGNLVTMEWWTHLWL 327
> CE24087
Length=781
Score = 64.3 bits (155), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 41/142 (28%), Positives = 72/142 (50%), Gaps = 4/142 (2%)
Query 21 VFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQM--HKLNWAMHSVKV 78
VF +YL A+ D + T + K + + +Y+ P+ M + + + +
Sbjct 76 VFEKSVKMSTYLLAVAVLDGYGYIKRLTRNTQKAIEVRLYA-PQDMLTGQSEFGLDTTIR 134
Query 79 SMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTSTDDDFERVL 138
++++ ED+F Y LD ++ + DF+ GAMEN GL F S LL + + ++ E +
Sbjct 135 ALEFFEDYFNISYPLDKIDLLALDDFSEGAMENWGLVTFRDSALLFNERKASVVAKEHIA 194
Query 139 AVVGHEYFHNWTGNRVTVRDWF 160
++ HE H W GN VT+ DW+
Sbjct 195 LIICHEIAHQWFGNLVTM-DWW 215
> YKL157w
Length=935
Score = 63.5 bits (153), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 46/147 (31%), Positives = 66/147 (44%), Gaps = 7/147 (4%)
Query 16 DRHFVVFTDPHPKPS-YLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWAMH 74
D V + PK S YL A + + + + + VY+ P +A
Sbjct 273 DGKKVTLFNTTPKMSTYLVAFIVAELK-----YVESKNFRIPVRVYATPGNEKHGQFAAD 327
Query 75 SVKVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTSTDDDF 134
++ + E FG +Y L + V +F+AGAMEN GL + LL ST D
Sbjct 328 LTAKTLAFFEKTFGIQYPLPKMDNVAVHEFSAGAMENWGLVTYRVVDLLLDKDNSTLDRI 387
Query 135 ERVLAVVGHEYFHNWTGNRVTVRDWFQ 161
+RV VV HE H W GN VT+ DW++
Sbjct 388 QRVAEVVQHELAHQWFGNLVTM-DWWE 413
> 7301751
Length=1017
Score = 63.5 bits (153), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 42/138 (30%), Positives = 65/138 (47%), Gaps = 9/138 (6%)
Query 22 FTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWAMHSVKVSMK 81
F + P +YL A DF I SG AV++ + + +A+ +
Sbjct 322 FAESLPMSTYLVAYAISDFTHI------SSGN---FAVWARADAIKSAEYALSVGPRILT 372
Query 82 WEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTSTDDDFERVLAVV 141
+ +D F + L ++ + +F AGAMEN GL F + +L +T ++ +RV +VV
Sbjct 373 FLQDFFNVTFPLPKIDMIALPEFQAGAMENWGLITFRETAMLYDPGVATANNKQRVASVV 432
Query 142 GHEYFHNWTGNRVTVRDW 159
GHE H W GN VT W
Sbjct 433 GHELAHQWFGNLVTPSWW 450
> 7301517
Length=1078
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 46/145 (31%), Positives = 75/145 (51%), Gaps = 9/145 (6%)
Query 27 PK-PSYLFAILAGDFAAIVDT--FTTKSGKEVALAVYSEPEQMHKLNWAMHSVKVSMKWE 83
PK P+YL A + + +VD+ + SG + +++ P+ + ++A V+ + +
Sbjct 374 PKMPTYLVAFIVSN---MVDSRLASQDSGLTPRVEIWTRPQFVGMTHYAYKMVRKFLPYY 430
Query 84 EDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLAS--MQTSTDDDFERVLA-V 140
ED FG + L ++ V DF AMEN GL F S LL +Q ++ + +V+A +
Sbjct 431 EDFFGIKNKLPKIDLVSVPDFGFAAMENWGLITFRDSALLVPEDLQLASSSEHMQVVAGI 490
Query 141 VGHEYFHNWTGNRVTVRDWFQLTLK 165
+ HE H W GN VT + W L LK
Sbjct 491 IAHELAHQWFGNLVTPKWWDDLWLK 515
> CE27507
Length=1073
Score = 62.4 bits (150), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 42/153 (27%), Positives = 69/153 (45%), Gaps = 14/153 (9%)
Query 21 VFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWAMHSVKVSM 80
VF P +YL A G+F + +++ + + + V++ PE + + + + V
Sbjct 298 VFQTTPPMSTYLLAFAIGEFVKL----ESRTERGIPVTVWTYPEDVMSMKFTLEYAPVIF 353
Query 81 KWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTSTDDDFE----- 135
ED Y L ++ +F+ G MEN GL +F + + A TD E
Sbjct 354 DRLEDALEIPYPLPKVDLIAARNFHVGGMENWGLVVFEFASI-AYTPPITDHVNETVDRM 412
Query 136 ----RVLAVVGHEYFHNWTGNRVTVRDWFQLTL 164
R+ ++ HE H W GN VT+RDW +L L
Sbjct 413 YNEFRIGKLIAHEAAHQWFGNLVTMRDWSELFL 445
> CE18905
Length=747
Score = 61.6 bits (148), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 39/130 (30%), Positives = 63/130 (48%), Gaps = 12/130 (9%)
Query 30 SYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWAMHSVKVSMKWEEDHFGR 89
SY+ A+ GD + T V + VY++P + +++ A++ ++ ++ E FG
Sbjct 227 SYILALFIGD----IQFKETILNNGVRIRVYTDPVNIDRVDHALNISRIVLEGFERQFGI 282
Query 90 EYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTSTDDDFERVLAVVGHEYFHNW 149
Y ++ + V +F GAMEN GL I N L+ D V +V HE H W
Sbjct 283 RYPMEKLDFVSVQNFKFGAMENWGLVIHNAYSLIG--------DPMDVTEIVIHEIAHQW 334
Query 150 TGNRVTVRDW 159
GN VT++ W
Sbjct 335 FGNLVTMKYW 344
> CE20255
Length=1045
Score = 60.8 bits (146), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 43/138 (31%), Positives = 63/138 (45%), Gaps = 4/138 (2%)
Query 27 PKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWAMHSVKVSMKWEEDH 86
P +YLFA D+ + +TF SG+ V VY +P ++ S+ + + ED+
Sbjct 346 PMSTYLFAFSVSDYPYL-ETF---SGRGVRSRVYCDPTKLVDAQLITKSIGPVLDFYEDY 401
Query 87 FGREYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTSTDDDFERVLAVVGHEYF 146
FG Y L+ +V V + AMEN GL + L + V +V HE
Sbjct 402 FGIPYPLEKLDVVIVPALSVTAMENWGLITIRQTNGLYTEGRFAISQKHDVQEIVAHELA 461
Query 147 HNWTGNRVTVRDWFQLTL 164
H W GN VT++ W L L
Sbjct 462 HQWFGNLVTMKWWNDLWL 479
> ECU01g1470
Length=864
Score = 59.7 bits (143), Expect = 3e-09, Method: Composition-based stats.
Identities = 45/154 (29%), Positives = 71/154 (46%), Gaps = 9/154 (5%)
Query 16 DRHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWAMHS 75
DR F + +YL A + G+ + I D +K G V L VY + ++ + +
Sbjct 197 DRKIEYFEETCKMSTYLVAFVVGELSYIEDW--SKDG--VRLRVYGDSSEVEWGRYGLEV 252
Query 76 VKVSMKWEEDHFGREYDLD-----TFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTST 130
K +++ ++FG Y+ ++ + +F++GAMEN GL F LL S
Sbjct 253 GKRCLEYFSEYFGVGYEFPRAGSAKIDMVGIPNFSSGAMENWGLITFRRESLLYVPGKSN 312
Query 131 DDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTL 164
+D + V V HE H W GN VT+ W L L
Sbjct 313 VEDMKNVAGTVCHELGHMWFGNLVTMSWWDDLWL 346
> ECU01g0140
Length=864
Score = 59.7 bits (143), Expect = 3e-09, Method: Composition-based stats.
Identities = 45/154 (29%), Positives = 71/154 (46%), Gaps = 9/154 (5%)
Query 16 DRHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWAMHS 75
DR F + +YL A + G+ + I D +K G V L VY + ++ + +
Sbjct 197 DRKIEYFEETCKMSTYLVAFVVGELSYIEDW--SKDG--VRLRVYGDSSEVEWGRYGLEV 252
Query 76 VKVSMKWEEDHFGREYDLD-----TFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTST 130
K +++ ++FG Y+ ++ + +F++GAMEN GL F LL S
Sbjct 253 GKRCLEYFSEYFGVGYEFPRAGSAKIDMVGIPNFSSGAMENWGLITFRRESLLYVPGKSN 312
Query 131 DDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTL 164
+D + V V HE H W GN VT+ W L L
Sbjct 313 VEDMKNVAGTVCHELGHMWFGNLVTMSWWDDLWL 346
> ECU08g0070
Length=864
Score = 58.9 bits (141), Expect = 4e-09, Method: Composition-based stats.
Identities = 45/154 (29%), Positives = 71/154 (46%), Gaps = 9/154 (5%)
Query 16 DRHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWAMHS 75
DR F + +YL A + G+ + I D +K G V L VY + ++ + +
Sbjct 197 DRKIEYFEETCKMSTYLVAFVVGELSYIEDW--SKDG--VRLRVYGDSSEVEWGRYGLEV 252
Query 76 VKVSMKWEEDHFGREYDLD-----TFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTST 130
K +++ ++FG Y+ ++ + +F++GAMEN GL F LL S
Sbjct 253 GKRCLEYFSEYFGVGYEFPRAGSAKIDMVGIPNFSSGAMENWGLITFRRESLLYVPGKSN 312
Query 131 DDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTL 164
+D + V V HE H W GN VT+ W L L
Sbjct 313 VEDMKNVAETVCHELGHMWFGNLVTMSWWDDLWL 346
> SPBC1921.05
Length=882
Score = 58.9 bits (141), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 50/160 (31%), Positives = 73/160 (45%), Gaps = 6/160 (3%)
Query 1 SNGNKTEEGPVQDSPDRHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTT-KSGKEVALAV 59
SN N EE V+D F + +YL A + + V+ FT K + + V
Sbjct 178 SNMNAVEE-TVKDG--LKTARFAETCRMSTYLLAWIVAELE-YVEYFTPGKHCPRLPVRV 233
Query 60 YSEPEQMHKLNWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNC 119
Y+ P + +A ++ + FG Y L ++ + DF AGAMEN GL +
Sbjct 234 YTTPGFSEQGKFAAELGAKTLDFFSGVFGEPYPLPKCDMVAIPDFEAGAMENWGLVTYRL 293
Query 120 SLLLASMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDW 159
+ +L S + S ERV VV HE H W GN VT++ W
Sbjct 294 AAILVS-EDSAATVIERVAEVVQHELAHQWFGNLVTMQFW 332
> 7291179
Length=931
Score = 58.5 bits (140), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 39/144 (27%), Positives = 64/144 (44%), Gaps = 2/144 (1%)
Query 22 FTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWAMHSVKVSMK 81
F + HP +YL A + F FT+ + ++ ++ P+ + + +A V +
Sbjct 214 FVESHPMQTYLVAFMISKFDR--PGFTSSERSDCPISTWARPDALSQTEFANMVVAPLLS 271
Query 82 WEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTSTDDDFERVLAVV 141
+ ED F Y ++ + DF + EN GL F LL Q S+ DD + V V
Sbjct 272 FYEDLFNSTYRQKKIDLVALPDFTFKSKENWGLPAFAEESLLYDSQRSSIDDQQGVARAV 331
Query 142 GHEYFHNWTGNRVTVRDWFQLTLK 165
+ W GN V+V W ++ LK
Sbjct 332 AMMVVNQWFGNLVSVAWWHEIWLK 355
> CE26001
Length=1815
Score = 56.2 bits (134), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 39/133 (29%), Positives = 66/133 (49%), Gaps = 7/133 (5%)
Query 30 SYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWAMHSVKVSMKWEEDHFGR 89
+YL A+ G F+ + T++G + +S EQ + A++ ++ + E++F
Sbjct 1205 TYLLALCVGHFSNLATV--TRTGVLTRVWTWSGMEQYGEF--ALNVTAGTIDFMENYFSY 1260
Query 90 EYDLDTFNVACVSDF--NAGAMENKGLNIFNCSLLLASMQTSTDDDFERVLAVVGHEYFH 147
++ L +V + ++ NAGAMEN GL I SL + +T D V HE H
Sbjct 1261 DFPLKKLDVMALPEYTMNAGAMENWGLIIGEYSLFMFDPDYATTRDITEVAETTAHEVVH 1320
Query 148 NWTGNRVTVRDWF 160
W G+ VT+ DW+
Sbjct 1321 QWFGDIVTL-DWW 1332
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 29/102 (28%), Positives = 45/102 (44%), Gaps = 0/102 (0%)
Query 63 PEQMHKLNWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCSLL 122
P L +A + + ++ G ++ L + + +F AGAMEN GL I+ +
Sbjct 310 PGTEQYLQFAAQNAGECLYQLGEYTGIKFPLSKADQLGMPEFLAGAMENWGLIIYKYQYI 369
Query 123 LASMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTL 164
+ T T + E V+ HE H W G+ VT W L L
Sbjct 370 AYNPTTMTTRNMEAAAKVMCHELAHQWFGDLVTTAWWDDLFL 411
> Hs7019561
Length=1024
Score = 56.2 bits (134), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 39/120 (32%), Positives = 60/120 (50%), Gaps = 4/120 (3%)
Query 48 TTKSGKEVALAVYSEPEQMHKL--NWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFN 105
TTKSG V + +Y+ P+ + + ++A+H K +++ ED+F Y L ++ V
Sbjct 345 TTKSG--VVVRLYARPDAIRRGSGDYALHITKRLIEFYEDYFKVPYSLPKLDLLAVPKHP 402
Query 106 AGAMENKGLNIFNCSLLLASMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTLK 165
AMEN GL+IF +L S+ V V+ HE H W G+ VT W + LK
Sbjct 403 YAAMENWGLSIFVEQRILLDPSVSSISYLLDVTMVIVHEICHQWFGDLVTPVWWEDVWLK 462
> Hs4505029
Length=611
Score = 54.7 bits (130), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 45/156 (28%), Positives = 61/156 (39%), Gaps = 25/156 (16%)
Query 8 EGPVQDSPDRHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMH 67
E P + P R F P P YL A++ G + + G V+SE EQ+
Sbjct 178 ETPDPEDPSRKIYKFIQKVPIPCYLIALVVGALE------SRQIGPRTL--VWSEKEQVE 229
Query 68 KLNWAMHSVKVSMKWEEDHFGR----EYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLL 123
K + + +K ED G +YDL + F G MEN L +LL
Sbjct 230 KSAYEFSETESMLKIAEDLGGPYVWGQYDL----LVLPPSFPYGGMENPCLTFVTPTLLA 285
Query 124 ASMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDW 159
S V+ HE H+WTGN VT + W
Sbjct 286 GDKSLSN---------VIAHEISHSWTGNLVTNKTW 312
> 7300769
Length=1646
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 45/158 (28%), Positives = 70/158 (44%), Gaps = 21/158 (13%)
Query 6 TEEGPVQDSPDRHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQ 65
TEEG D V+ +YL A + +F A K+ VY+ PE
Sbjct 228 TEEGIFSD-------VYKTTPKMSTYLLAFIISEFVA---------RKDDDFGVYARPEY 271
Query 66 MHKLNWAMHSVKVSMKWEEDHFGRE--YDL--DTFNVACVSDFNAGAMENKGLNIFNCSL 121
+ + ++V + + E + + Y + D ++A + DF+AGAMEN GL +
Sbjct 272 YAQTQYP-YNVGIQILEEMGQYLDKDYYSMGNDKMDMAAIPDFSAGAMENWGLLTYRERS 330
Query 122 LLASMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDW 159
LL +T + + AVV HE H W G+ VT + W
Sbjct 331 LLVDESATTLASRQSIAAVVAHEQAHMWFGDLVTCKWW 368
Score = 41.6 bits (96), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 40/150 (26%), Positives = 61/150 (40%), Gaps = 12/150 (8%)
Query 14 SPDRHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWAM 73
S DR F + +YL A + +++A G E AV + PE ++
Sbjct 1161 SQDRFVDHFKETPVMSTYLLAFMVANYSA--------RGNESEFAVLTRPEFYDNTEFSY 1212
Query 74 HSVKVSMKWEEDHFGREY-DL--DTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTST 130
H + + + F Y +L D A F MEN GL I++ +L+ S
Sbjct 1213 HVGQQVVSAYGELFQSPYAELGNDVLQYASSPRFPHNGMENWGLIIYSDDVLIQEPGYSD 1272
Query 131 D-DDFERVLAVVGHEYFHNWTGNRVTVRDW 159
D D E + ++ HE H W G+ VT W
Sbjct 1273 DWSDKEFAIRIIAHETSHMWFGDSVTFSWW 1302
> 7300767
Length=988
Score = 51.2 bits (121), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 37/135 (27%), Positives = 62/135 (45%), Gaps = 10/135 (7%)
Query 27 PKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWAMHSVKVSMKWEEDH 86
P +YL A + DF +I +T+ + + ++Y+ P K A+ + ++ ED+
Sbjct 224 PISTYLVAFVISDFGSISETY-----RGITQSIYTSPTSKEKGQVALKNAVRTVAALEDY 278
Query 87 FGREYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTSTDDDFERVLAVV--GHE 144
FG Y L + + AMEN GL + LL ++ + D +R L ++ HE
Sbjct 279 FGVSYPLPKLDHVALKKNYGAAMENWGLITYKDVNLLKNI---SSDGQKRKLDLITQNHE 335
Query 145 YFHNWTGNRVTVRDW 159
H W GN V+ W
Sbjct 336 IAHQWFGNLVSPEWW 350
> CE24840
Length=609
Score = 49.7 bits (117), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 44/149 (29%), Positives = 58/149 (38%), Gaps = 29/149 (19%)
Query 17 RHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVA--LAVYSEPEQMHKLNWAMH 74
R F P PSYL AI+ G KE++ AV++EP Q +
Sbjct 188 RTIFSFKQPVSIPSYLLAIVVGHL----------ERKEISERCAVWAEPSQAEASFYEFA 237
Query 75 SVKVSMKWEEDHFGR----EYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTST 130
+ +K ED G YDL V + F G MEN L +LL
Sbjct 238 ETEKILKVAEDVAGPYVWGRYDL----VVLPATFPFGGMENPCLTFITPTLLAGD----- 288
Query 131 DDDFERVLAVVGHEYFHNWTGNRVTVRDW 159
++ V+ HE H+WTGN VT W
Sbjct 289 ----RSLVNVIAHEISHSWTGNLVTNFSW 313
> CE28160
Length=625
Score = 48.5 bits (114), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 36/139 (25%), Positives = 58/139 (41%), Gaps = 19/139 (13%)
Query 22 FTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWAMHSVKVSMK 81
+ P PSYL AI+ G K AV++EP + K +W + +
Sbjct 195 YKQPVAIPSYLIAIVVGCLE--------KRDISDRCAVWAEPSVVDKAHWEFAETEDILA 246
Query 82 WEEDHFGREYDLDTFNVACVS-DFNAGAMENKGLNIFNCSLLLASMQTSTDDDFERVLAV 140
E+ G+ Y +++ C+ F G MEN L +L+ +++V
Sbjct 247 SAEEIAGK-YIWGRYDMVCLPPSFPFGGMENPCLTFLTPTLIAGD---------RSLVSV 296
Query 141 VGHEYFHNWTGNRVTVRDW 159
+ HE H+W+GN VT W
Sbjct 297 IAHEIAHSWSGNLVTNSSW 315
> SPCC1322.05c
Length=612
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 42/144 (29%), Positives = 62/144 (43%), Gaps = 26/144 (18%)
Query 21 VFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWAM-HSVKVS 79
+F +P PSYLF IL+GD A +T G + VY+EP + + H ++
Sbjct 188 LFEQKNPIPSYLFCILSGDLA------STNIGPRSS--VYTEPGNLLACKYEFEHDMENF 239
Query 80 MKWEED----HFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTSTDDDFE 135
M+ E + YD V F G MEN F L+A +++ +
Sbjct 240 MEAAEQLTLPYCWTRYDF----VILPPSFPYGGMENPNAT-FATPTLIAGDRSNVN---- 290
Query 136 RVLAVVGHEYFHNWTGNRVTVRDW 159
V+ HE H+W+GN VT W
Sbjct 291 ----VIAHELAHSWSGNLVTNESW 310
> YNL045w
Length=671
Score = 45.4 bits (106), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 38/148 (25%), Positives = 63/148 (42%), Gaps = 20/148 (13%)
Query 14 SPDRHFVVFTDPHPKPSYLFAILAGDFA-AIVDTFTTKSGKEVALAVYSEPEQMHKLNWA 72
S D + F P P+YL I +GD + A + +T VY+EP ++ W
Sbjct 227 SKDTNIYRFEQKVPIPAYLIGIASGDLSSAPIGPRST---------VYTEPFRLKDCQWE 277
Query 73 MHSVKVSMKWEEDHFGREYDLDTFNVAC-VSDFNAGAMENKGLNIFNCSLLLASMQTSTD 131
+ + EY+ T+++ V + G ME+ + F L+A +++ D
Sbjct 278 FENDVEKFIQTAEKIIFEYEWGTYDILVNVDSYPYGGMESPNMT-FATPTLIAHDRSNID 336
Query 132 DDFERVLAVVGHEYFHNWTGNRVTVRDW 159
V+ HE H+W+GN VT W
Sbjct 337 --------VIAHELAHSWSGNLVTNCSW 356
> YIL137c
Length=946
Score = 45.1 bits (105), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 31/151 (20%), Positives = 62/151 (41%), Gaps = 2/151 (1%)
Query 13 DSPDRHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWA 72
DS +H V F + +F GD + + + + +++Y+ P + +
Sbjct 200 DSSQKHLVKFAKTPLMTTSVFGFSIGDLEFLKTEIKLEGDRTIPVSIYA-PWDIANAAFT 258
Query 73 MHSVKVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCS-LLLASMQTSTD 131
+ +V+ + E +F Y L + + + AMEN G+ + LL+ + +
Sbjct 259 LDTVQKYLPLLESYFKCPYPLPKLDFVLLPYLSDMAMENFGMITIQLNHLLIPPNALANE 318
Query 132 DDFERVLAVVGHEYFHNWTGNRVTVRDWFQL 162
E+ ++ HE H W GN ++ W L
Sbjct 319 TVREQAQQLIVHELVHQWMGNYISFDSWESL 349
> 7291558
Length=844
Score = 39.3 bits (90), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 35/136 (25%), Positives = 53/136 (38%), Gaps = 1/136 (0%)
Query 30 SYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWAMHSVKVSMKWEEDHFGR 89
+YL A D T +K+ V + +P+ + + +M + + E+ F
Sbjct 156 TYLVAFAVHDLENAA-TEESKTSNRVIFRNWMQPKLLGQEMISMEIAPKLLSFYENLFQI 214
Query 90 EYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTSTDDDFERVLAVVGHEYFHNW 149
+ L + V AMEN GL +N L + + V HEY H W
Sbjct 215 NFPLAKVDQLTVPTHRFTAMENWGLVTYNEERLPQNQGDYPQKQKDSTAFTVAHEYAHQW 274
Query 150 TGNRVTVRDWFQLTLK 165
GN VT+ W L LK
Sbjct 275 FGNLVTMNWWNDLWLK 290
> At4g33090
Length=873
Score = 38.1 bits (87), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 21/64 (32%), Positives = 30/64 (46%), Gaps = 0/64 (0%)
Query 86 HFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTSTDDDFERVLAVVGHEY 145
+F Y L ++ + DF AGAMEN GL + + LL Q S + +RV +
Sbjct 266 YFAVPYPLPKMDMIAIPDFAAGAMENYGLVTYRETALLYDEQHSAASNKQRVSYLATDSL 325
Query 146 FHNW 149
F W
Sbjct 326 FPEW 329
> Hs20070296
Length=494
Score = 37.7 bits (86), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 39/140 (27%), Positives = 58/140 (41%), Gaps = 18/140 (12%)
Query 26 HPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWAMHS-VKVSMKWEE 84
HP P+YL A++AGD ++ E L P KL+ A+ + + +
Sbjct 21 HPVPAYLVALVAGDLKPADIGPRSRVWAEPCLL----PTATSKLSGAVEQWLSAAERLYG 76
Query 85 DHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTSTDDDFERVLAVVGHE 144
+ YD+ V F AMEN L S+L D+F ++ V HE
Sbjct 77 PYMWGRYDI----VFLPPSFPIVAMENPCLTFIISSIL-------ESDEF--LVIDVIHE 123
Query 145 YFHNWTGNRVTVRDWFQLTL 164
H+W GN VT W ++ L
Sbjct 124 VAHSWFGNAVTNATWEEMWL 143
> At5g13520
Length=616
Score = 37.4 bits (85), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 40/146 (27%), Positives = 59/146 (40%), Gaps = 26/146 (17%)
Query 16 DRHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVA--LAVYSEPEQMHKLNWA- 72
DR F P P YLFA G+ +EV VY+E + L+ A
Sbjct 196 DRVVEEFAMEQPIPPYLFAFAVGELGF----------REVGPRTRVYTESAAIEVLDAAA 245
Query 73 --MHSVKVSMKWEEDHFGREYDLDTFNVACVS-DFNAGAMENKGLNIFNCSLLLASMQTS 129
+ +K E FG +Y+ + F++ + F G MEN + +F ++ T
Sbjct 246 LEFAGTEDMIKQGEKLFG-DYEWERFDLLVLPPSFPYGGMENPRM-VFLTPTVIKGDATG 303
Query 130 TDDDFERVLAVVGHEYFHNWTGNRVT 155
VV HE H+WTGN +T
Sbjct 304 AQ--------VVAHELAHSWTGNLIT 321
> Hs13443031
Length=657
Score = 35.4 bits (80), Expect = 0.055, Method: Compositional matrix adjust.
Identities = 42/157 (26%), Positives = 62/157 (39%), Gaps = 27/157 (17%)
Query 12 QDSPDRHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNW 71
+ P++ F F P PSYL A+ GD + ++ V++EP +
Sbjct 220 KRGPNKFF--FQMCQPIPSYLIALAIGDLVSAEVGPRSR--------VWAEPCLIDAAKE 269
Query 72 AMHSVKVSMKWEEDHFGR----EYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQ 127
++ + E FG YDL + F G MEN L F LLA +
Sbjct 270 YNGVIEEFLATGEKLFGPYVWGRYDL----LFMPPSFPFGGMENPCLT-FVTPCLLAGDR 324
Query 128 TSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTL 164
+ D V+ HE H+W GN VT +W + L
Sbjct 325 SLAD--------VIIHEISHSWFGNLVTNANWGEFWL 353
> 7298498
Length=613
Score = 35.0 bits (79), Expect = 0.072, Method: Compositional matrix adjust.
Identities = 41/153 (26%), Positives = 58/153 (37%), Gaps = 45/153 (29%)
Query 21 VFTDPHPKPSYLFAILAGDFA--------------AIVDTFTTKSGKEVALAVYSEPEQM 66
+F P P+YL AI G AIVD + +SE M
Sbjct 188 LFKQEVPIPAYLVAIAIGKLVSRPLGENSSVWAEEAIVDACAEE---------FSETATM 238
Query 67 HKLNWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASM 126
+K + + + ++YDL + F G MEN L F LLA
Sbjct 239 ---------LKTATELCGPYVWKQYDL----LVMPPSFPFGGMENPCLT-FVTPTLLAGD 284
Query 127 QTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDW 159
++ D VV HE H+WTGN VT +++
Sbjct 285 KSLAD--------VVAHEIAHSWTGNLVTNKNF 309
> CE13593
Length=556
Score = 33.9 bits (76), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 18/59 (30%), Positives = 28/59 (47%), Gaps = 7/59 (11%)
Query 107 GAMENKGLNIFNCSLLLASMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTLK 165
G MEN G + S +T D ++ ++ HE H+W GNR T+ W + L+
Sbjct 187 GGMENWGH-------ITVSETLATSGDDAHLIYLIAHEIAHHWIGNRATIDSWNWICLQ 238
Lambda K H
0.319 0.133 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2353551590
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40