bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0532_orf2
Length=142
Score E
Sequences producing significant alignments: (Bits) Value
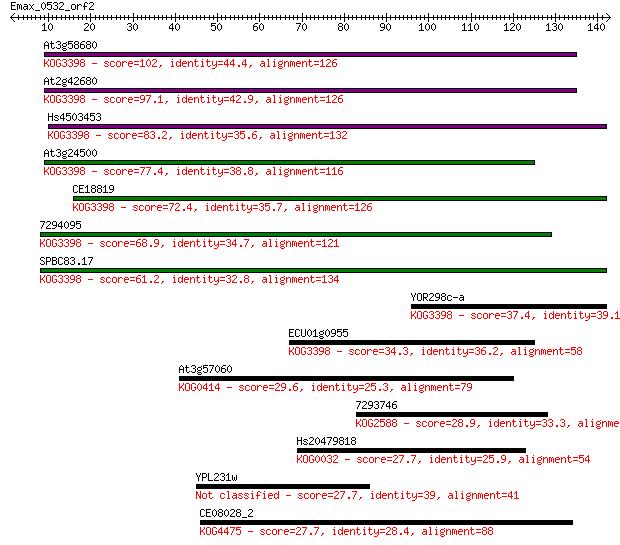
At3g58680 102 2e-22
At2g42680 97.1 9e-21
Hs4503453 83.2 2e-16
At3g24500 77.4 7e-15
CE18819 72.4 3e-13
7294095 68.9 3e-12
SPBC83.17 61.2 7e-10
YOR298c-a 37.4 0.010
ECU01g0955 34.3 0.073
At3g57060 29.6 1.9
7293746 28.9 3.7
Hs20479818 27.7 6.4
YPL231w 27.7 6.7
CE08028_2 27.7 7.7
> At3g58680
Length=142
Score = 102 bits (254), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 56/131 (42%), Positives = 75/131 (57%), Gaps = 5/131 (3%)
Query 9 QDWTPVTFQKTSPGGRRVVKEQEVNKTRRAGEVIETEKKFLGGRNKTTKANLIANAKKVE 68
QDW PV +K +P E+ VN RR+G IET +KF G NK + N KK++
Sbjct 9 QDWEPVVIRKRAPNAAAKRDEKTVNAARRSGADIETVRKFNAGSNKAASSGTSLNTKKLD 68
Query 69 EDTGDYHVDRVSTDFSRALAQARRNKGMTQAQLAQAINEKPSVVSEYESGKHLDGE---- 124
+DT + DRV T+ +A+ QAR K +TQ+QLA INEKP V+ EYESGK + +
Sbjct 69 DDTENLSHDRVPTELKKAIMQARGEKKLTQSQLAHLINEKPQVIQEYESGKAIPNQQILS 128
Query 125 -LHRGTGRFLK 134
L R G L+
Sbjct 129 KLERALGAKLR 139
> At2g42680
Length=142
Score = 97.1 bits (240), Expect = 9e-21, Method: Compositional matrix adjust.
Identities = 54/131 (41%), Positives = 74/131 (56%), Gaps = 5/131 (3%)
Query 9 QDWTPVTFQKTSPGGRRVVKEQEVNKTRRAGEVIETEKKFLGGRNKTTKANLIANAKKVE 68
QDW PV +K E+ VN RR+G IET +KF G NK + N K ++
Sbjct 9 QDWEPVVIRKKPANAAAKRDEKTVNAARRSGADIETVRKFNAGTNKAASSGTSLNTKMLD 68
Query 69 EDTGDYHVDRVSTDFSRALAQARRNKGMTQAQLAQAINEKPSVVSEYESGKHLDGE---- 124
+DT + +RV T+ +A+ QAR +K +TQ+QLAQ INEKP V+ EYESGK + +
Sbjct 69 DDTENLTHERVPTELKKAIMQARTDKKLTQSQLAQIINEKPQVIQEYESGKAIPNQQILS 128
Query 125 -LHRGTGRFLK 134
L R G L+
Sbjct 129 KLERALGAKLR 139
> Hs4503453
Length=148
Score = 83.2 bits (204), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 47/138 (34%), Positives = 78/138 (56%), Gaps = 8/138 (5%)
Query 10 DWTPVT-FQKTSPGGRRVVKEQEVNKTRRAGEVIETEKKFLGGRNKTTKANLIANAKKVE 68
DW VT +K P + +Q + +R GE +ET KK+ G+NK + ++ N K++
Sbjct 5 DWDTVTVLRKKGPTAAQAKSKQAILAAQRRGEDVETSKKWAAGQNK--QHSITKNTAKLD 62
Query 69 EDTGDYHVDRVSTDFSRALAQARRNKGMTQAQLAQAINEKPSVVSEYESGKHLD-----G 123
+T + H DRV+ + + + Q R++KG+TQ LA INEKP V+++YESG+ + G
Sbjct 63 RETEELHHDRVTLEVGKVIQQGRQSKGLTQKDLATKINEKPQVIADYESGRAIPNNQVLG 122
Query 124 ELHRGTGRFLKADSLQTP 141
++ R G L+ + P
Sbjct 123 KIERAIGLKLRGKDIGKP 140
> At3g24500
Length=148
Score = 77.4 bits (189), Expect = 7e-15, Method: Compositional matrix adjust.
Identities = 45/118 (38%), Positives = 65/118 (55%), Gaps = 2/118 (1%)
Query 9 QDWTPVTFQKTSPGGRRVVKEQEVNKTRRAGEVIETEKKFLGGRNKTTKANLIA--NAKK 66
QDW PV K+ + + + VN R G ++T KKF G NK K+ + N KK
Sbjct 11 QDWEPVVLHKSKQKSQDLRDPKAVNAALRNGVAVQTVKKFDAGSNKKGKSTAVPVINTKK 70
Query 67 VEEDTGDYHVDRVSTDFSRALAQARRNKGMTQAQLAQAINEKPSVVSEYESGKHLDGE 124
+EE+T +DRV + + +AR K M+QA LA+ INE+ VV EYE+GK + +
Sbjct 71 LEEETEPAAMDRVKAEVRLMIQKARLEKKMSQADLAKQINERTQVVQEYENGKAVPNQ 128
> CE18819
Length=156
Score = 72.4 bits (176), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 45/131 (34%), Positives = 67/131 (51%), Gaps = 7/131 (5%)
Query 16 FQKTSPGGRRVVKEQEVNKTRRAGEVIETEKKFLGGRNKTTKANLIANAKKVEEDTGDYH 75
K P + + ++N +RAG I TEKK + G N+ AN N +++E+T + H
Sbjct 19 ITKRGPVNKTLKSAAQLNAAQRAGVDISTEKKTMSGGNRQHSAN--KNTLRLDEETEELH 76
Query 76 VDRVSTDFSRALAQARRNKGMTQAQLAQAINEKPSVVSEYESGK-----HLDGELHRGTG 130
+V+ + + QAR KG TQ L+ INEKP VV EYESGK + ++ R G
Sbjct 77 HQKVALSLGKVMQQARATKGWTQKDLSTQINEKPQVVGEYESGKAVPNQQIMAKMERALG 136
Query 131 RFLKADSLQTP 141
L+ + P
Sbjct 137 VKLRGKDIGMP 147
> 7294095
Length=145
Score = 68.9 bits (167), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 42/125 (33%), Positives = 68/125 (54%), Gaps = 9/125 (7%)
Query 8 FQDWTPVT-FQKTSPGGRRVVKEQEVNKTRRAGEVIETEKKFLGGRNK---TTKANLIAN 63
DW VT +K +P + E VN+ RR G ++T++K+ G NK TTK N
Sbjct 1 MSDWDSVTVLRKKAPKSSTLKTESAVNQARRQGVAVDTQQKYGAGTNKQHVTTK-----N 55
Query 64 AKKVEEDTGDYHVDRVSTDFSRALAQARRNKGMTQAQLAQAINEKPSVVSEYESGKHLDG 123
K++ +T + D++ D + + Q R++KG++Q LA I EK VV++YE+G+ +
Sbjct 56 TAKLDRETEELRHDKIPLDVGKLIQQGRQSKGLSQKDLATKICEKQQVVTDYEAGRGIPN 115
Query 124 ELHRG 128
L G
Sbjct 116 NLILG 120
> SPBC83.17
Length=148
Score = 61.2 bits (147), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 44/143 (30%), Positives = 69/143 (48%), Gaps = 11/143 (7%)
Query 8 FQDWTPVT--FQKTSPGGR-RVVKEQ-EVNKTRRAGEVIETEKKFLGGRNKTTKANLIAN 63
DW VT + PG R V K Q ++N RRAG ++ TEKK+ G A +
Sbjct 1 MSDWDTVTKIGSRAGPGARTHVAKTQSQINSARRAGAIVGTEKKYATGNKSQDPAG--QH 58
Query 64 AKKVEEDTGDYHVDRVSTDFSRALAQARRNKGMTQAQLAQAINEKPSVVSEYESGKHLDG 123
K++ + ++A+ + R+ KG Q L+Q INEKP VV++YESG+ +
Sbjct 59 LTKIDRENEVKPPSTTGRSVAQAIQKGRQAKGWAQKDLSQRINEKPQVVNDYESGRAIPN 118
Query 124 E-----LHRGTGRFLKADSLQTP 141
+ + R G L+ ++ P
Sbjct 119 QQVLSKMERALGIKLRGQNIGAP 141
> YOR298c-a
Length=58
Score = 37.4 bits (85), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 18/51 (35%), Positives = 31/51 (60%), Gaps = 5/51 (9%)
Query 96 MTQAQLAQAINEKPSVVSEYESGKHLDGE-----LHRGTGRFLKADSLQTP 141
M+Q LA INEKP+VV++YE+ + + + L R G L+ +++ +P
Sbjct 1 MSQKDLATKINEKPTVVNDYEAARAIPNQQVLSKLERALGVKLRGNNIGSP 51
> ECU01g0955
Length=95
Score = 34.3 bits (77), Expect = 0.073, Method: Compositional matrix adjust.
Identities = 21/58 (36%), Positives = 32/58 (55%), Gaps = 3/58 (5%)
Query 67 VEEDTGDYHVDRVSTDFSRALAQARRNKGMTQAQLAQAINEKPSVVSEYESGKHLDGE 124
V EDTG + VS A+A AR KGM++ LAQ + + S++ +E G+ + E
Sbjct 24 VPEDTG---LRIVSKRVGDAIANARAQKGMSRKDLAQKMKKNVSIIDSWERGEAVYNE 78
> At3g57060
Length=1439
Score = 29.6 bits (65), Expect = 1.9, Method: Composition-based stats.
Identities = 20/79 (25%), Positives = 33/79 (41%), Gaps = 0/79 (0%)
Query 41 VIETEKKFLGGRNKTTKANLIANAKKVEEDTGDYHVDRVSTDFSRALAQARRNKGMTQAQ 100
V E + + K K L A ++ EE +H+++ + + A+ R K T
Sbjct 1199 VTENFRSIINKGKKFAKPELKACIEEFEEKINKFHMEKKEQEETARNAEVHREKTKTMES 1258
Query 101 LAQAINEKPSVVSEYESGK 119
LA K V EY+ G+
Sbjct 1259 LAVLSKVKEEPVEEYDEGE 1277
> 7293746
Length=1113
Score = 28.9 bits (63), Expect = 3.7, Method: Composition-based stats.
Identities = 15/47 (31%), Positives = 27/47 (57%), Gaps = 2/47 (4%)
Query 83 FSRALAQARRNKGMTQA--QLAQAINEKPSVVSEYESGKHLDGELHR 127
++ L + R + TQ+ Q Q++ ++P V SE+ S H+ ELH+
Sbjct 53 YNMLLDEPRTHTQQTQSVDQQPQSVEQQPHVKSEHSSPVHIKEELHQ 99
> Hs20479818
Length=514
Score = 27.7 bits (60), Expect = 6.4, Method: Composition-based stats.
Identities = 14/54 (25%), Positives = 27/54 (50%), Gaps = 0/54 (0%)
Query 69 EDTGDYHVDRVSTDFSRALAQARRNKGMTQAQLAQAINEKPSVVSEYESGKHLD 122
+D + +++ TDF A+ + R++ M QA I P V+S ++ + D
Sbjct 252 DDNNEINLNIKVTDFGLAVKKQSRSEAMLQATCGTPIYMAPEVISAHDYSQQCD 305
> YPL231w
Length=1887
Score = 27.7 bits (60), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 16/41 (39%), Positives = 24/41 (58%), Gaps = 2/41 (4%)
Query 45 EKKFLGGRNKTTKANLIANAKKVEEDTGDYHVDRVSTDFSR 85
E+KFL + K T A L A + + G++ V+ V+T FSR
Sbjct 358 ERKFL--KEKDTVAELQAQLDYLNAELGEFFVNGVATSFSR 396
> CE08028_2
Length=6284
Score = 27.7 bits (60), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 25/110 (22%), Positives = 47/110 (42%), Gaps = 27/110 (24%)
Query 46 KKFLGGRNKTTKANLIANAK-------KVEEDTGDYHVD------------RVSTDFSRA 86
K+ G KT K I + K V EDTGDY V+ +++ +
Sbjct 4277 KELPGAAAKTIKIQKIDDNKYVLEIPSSVVEDTGDYKVEVANEAGSANSSGKITVEPKIT 4336
Query 87 LAQARRNKGMTQ---AQLAQAINEKPSVVSEYESGKHLDGELHRGTGRFL 133
+ +++ +T+ A+ + N KP +V Y++G+ + + RF+
Sbjct 4337 FLKPLKDQSITEGENAEFSVETNTKPRIVKWYKNGQEI-----KPNSRFI 4381
Lambda K H
0.312 0.128 0.358
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1603110344
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40