bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0528_orf4
Length=295
Score E
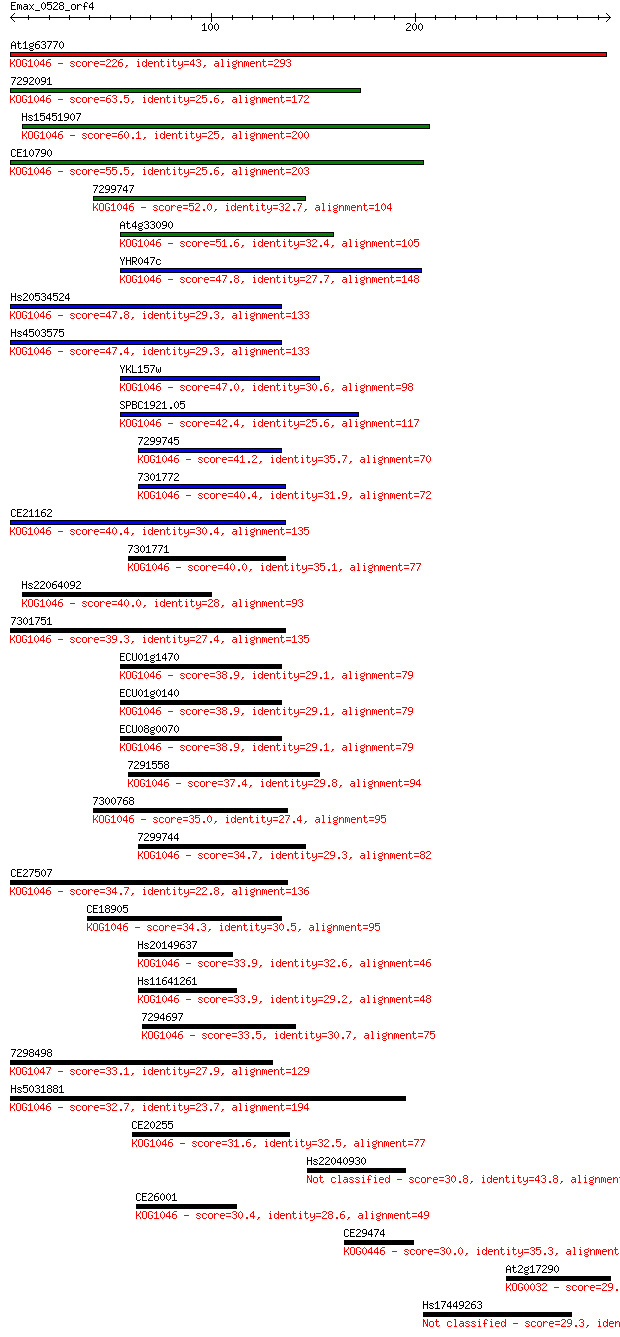
Sequences producing significant alignments: (Bits) Value
At1g63770 226 5e-59
7292091 63.5 5e-10
Hs15451907 60.1 5e-09
CE10790 55.5 1e-07
7299747 52.0 1e-06
At4g33090 51.6 2e-06
YHR047c 47.8 3e-05
Hs20534524 47.8 3e-05
Hs4503575 47.4 3e-05
YKL157w 47.0 5e-05
SPBC1921.05 42.4 0.001
7299745 41.2 0.003
7301772 40.4 0.005
CE21162 40.4 0.005
7301771 40.0 0.006
Hs22064092 40.0 0.006
7301751 39.3 0.010
ECU01g1470 38.9 0.012
ECU01g0140 38.9 0.012
ECU08g0070 38.9 0.013
7291558 37.4 0.035
7300768 35.0 0.21
7299744 34.7 0.22
CE27507 34.7 0.23
CE18905 34.3 0.29
Hs20149637 33.9 0.37
Hs11641261 33.9 0.39
7294697 33.5 0.58
7298498 33.1 0.66
Hs5031881 32.7 0.99
CE20255 31.6 1.8
Hs22040930 30.8 3.3
CE26001 30.4 4.1
CE29474 30.0 6.3
At2g17290 29.6 8.8
Hs17449263 29.3 9.1
> At1g63770
Length=964
Score = 226 bits (576), Expect = 5e-59, Method: Compositional matrix adjust.
Identities = 126/310 (40%), Positives = 180/310 (58%), Gaps = 23/310 (7%)
Query 1 LKEGLTVFREQLFMGTFMSAGVQRIKEVADLISRQFPEDDGPMAHAIRPESYLAMDN--- 57
LKEGLTVFR+Q F S V+RI +V+ L QFP+D GPMAH +RP SY+ +
Sbjct 399 LKEGLTVFRDQEFSSDMGSRTVKRIADVSKLRIYQFPQDAGPMAHPVRPHSYIKVYEKVW 458
Query 58 -FYTATVYDKGAEVVRIYYTLLGASGFRKGMDLYFKRHDNKPATCDDFRAAMADANGINL 116
F + + GAEVVR+Y TLLG GFRKG+DLYF+RHD + TC+DF AAM DAN +
Sbjct 459 LFTNSVLLYAGAEVVRMYKTLLGTQGFRKGIDLYFERHDEQAVTCEDFFAAMRDANNADF 518
Query 117 DQMDRWYSQAGTPRLEVLRSEYKEDEKAFEISFRQHTPPTRGQETKRPQLIPIKIGLIGR 176
+WYSQAGTP ++V+ S Y D + F + F Q PPT GQ TK P IP+ +GL+
Sbjct 519 ANFLQWYSQAGTPVVKVV-SSYNADARTFSLKFSQEIPPTPGQPTKEPTFIPVVVGLLDS 577
Query 177 AS--------HQD-----LLQPAVVLEMTEEEQTFRVRGVREDCVPSILRGFSAPVRLIN 223
+ H D + + +L +T++E+ F + E VPS+ RGFSAPVR +
Sbjct 578 SGKDITLSSVHHDGTVQTISGSSTILRVTKKEEEFVFSDIPERPVPSLFRGFSAPVR-VE 636
Query 224 RPQTAKESAFLLAYDTDPVTKWFASRALATPIVISRSKRISAKGNAQKLEQLPDDYVEGL 283
+ + FLLA+D+D +W A + LA ++++ + + K L +V+GL
Sbjct 637 TDLSNDDLFFLLAHDSDEFNRWEAGQVLARKLMLN----LVSDFQQNKPLALNPKFVQGL 692
Query 284 RAIIYDQATD 293
+++ D + D
Sbjct 693 GSVLSDSSLD 702
> 7292091
Length=866
Score = 63.5 bits (153), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 44/175 (25%), Positives = 76/175 (43%), Gaps = 3/175 (1%)
Query 1 LKEGLTVFREQLFMGTFMSAGVQRIKEVADLISRQFPEDDGPMAHAIR-PESYLA-MDNF 58
L EG F E L + + V D+ +R D +H I P + + +D
Sbjct 327 LNEGYASFVEFLCVHHLFPEYDIWTQFVTDMYTRALELDSLKNSHPIEVPVGHPSEIDEI 386
Query 59 YTATVYDKGAEVVRIYYTLLGASGFRKGMDLYFKRHDNKPATCDDFRAAMADANGINL-D 117
+ Y+KGA V+R+ + +G FRKGM+LY RH +D AA+ +A+ N+ D
Sbjct 387 FDEISYNKGASVIRMLHDYIGEDTFRKGMNLYLTRHQYGNTCTEDLWAALQEASSKNVSD 446
Query 118 QMDRWYSQAGTPRLEVLRSEYKEDEKAFEISFRQHTPPTRGQETKRPQLIPIKIG 172
M W G P + V + ++++ + + T + ++PI +
Sbjct 447 VMTSWTQHKGFPVVSVESEQTSKNQRLLRLKQCKFTADGSQADENCLWVVPISVS 501
> Hs15451907
Length=875
Score = 60.1 bits (144), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 50/202 (24%), Positives = 93/202 (46%), Gaps = 16/202 (7%)
Query 7 VFREQLFMGTFMSAGVQRIKEVADLISRQFPEDDGPMAHAIRPESYLAMDNFYTATVYDK 66
F E F+SA R +E+ L ++ P+ ++ S +D + A Y K
Sbjct 345 CFPEYDIWTQFVSADYTRAQELDAL------DNSHPIEVSVGHPS--EVDEIFDAISYSK 396
Query 67 GAEVVRIYYTLLGASGFRKGMDLYFKRHDNKPATCDDFRAAMADANGINLDQ-MDRWYSQ 125
GA V+R+ + +G F+KGM++Y + K A +D ++ +A+G + M+ W Q
Sbjct 397 GASVIRMLHDYIGDKDFKKGMNMYLTKFQQKNAATEDLWESLENASGKPIAAVMNTWTKQ 456
Query 126 AGTPRLEVLRSEYKEDEKAFEISFRQHTPPTRGQETKRPQ-LIPIKIGLIGRASHQDLLQ 184
G P + V +E ED++ +S ++ PQ ++PI I ++ +D Q
Sbjct 457 MGFPLIYV-EAEQVEDDRLLRLSQKKFCAGGSYVGEDCPQWMVPITI-----STSEDPNQ 510
Query 185 PAVVLEMTEEEQTFRVRGVRED 206
+ + M + E ++ V+ D
Sbjct 511 AKLKILMDKPEMNVVLKNVKPD 532
> CE10790
Length=884
Score = 55.5 bits (132), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 52/206 (25%), Positives = 87/206 (42%), Gaps = 9/206 (4%)
Query 1 LKEGLTVFREQLFMGTFMSAGVQRIKEVADLISRQFPEDDGPMAHAIRPE--SYLAMDNF 58
LKEG F E +F+G + + D ++ D +H I E + +D
Sbjct 334 LKEGFASFMEYMFVGANCPEFKIWLHFLNDELASGMGLDALRNSHPIEVEIDNPNELDEI 393
Query 59 YTATVYDKGAEVVRIYYTLLGASGFRKGMDLYFKRHDNKPATCDDFRAAMADANGINLDQ 118
Y + Y K V R+ L F+KG+ LY KR A D A+++A+G N+++
Sbjct 394 YDSITYAKSNSVNRMLCYYLSEPVFQKGLRLYLKRFQYSNAVTQDLWTALSEASGQNVNE 453
Query 119 -MDRWYSQAGTPRLEVLRSEYKEDEKAFEISFRQHTPPTRGQETKRPQLIPIKIGLIGRA 177
M W Q G P VL+ ++D ++ Q + G E + + I + +
Sbjct 454 LMSGWTQQMGFP---VLKVSQRQDGNNRILTVEQRRFISDGGEDPKNSQWQVPITVAVGS 510
Query 178 SHQDLLQPAVVLEMTEEEQTFRVRGV 203
S D+ + E++Q F + GV
Sbjct 511 SPSDV---KARFLLKEKQQEFTIEGV 533
> 7299747
Length=985
Score = 52.0 bits (123), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 34/109 (31%), Positives = 50/109 (45%), Gaps = 7/109 (6%)
Query 42 PMAHAIRPESYLAMDNFYTATVYDKGAEVVRIYYTLLGASGFRKGMDLYFKRHDNKPATC 101
P+ +I ES + ++ Y KGA +VR+ L+G R Y RH AT
Sbjct 514 PIVKSI--ESPAEITEYFDTITYSKGAALVRMLENLVGEEKLRNATTRYLVRHIYSTATT 571
Query 102 DDFRAAMADANGINLDQ---MDRWYSQAGTPRLEVLR--SEYKEDEKAF 145
+D+ A+ + G+ D M W Q G P +EV + S YK +K F
Sbjct 572 EDYLTAVEEEEGLEFDVKQIMQTWTEQMGLPVVEVEKSGSTYKLTQKRF 620
> At4g33090
Length=873
Score = 51.6 bits (122), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 34/107 (31%), Positives = 52/107 (48%), Gaps = 5/107 (4%)
Query 55 MDNFYTATVYDKGAEVVRIYYTLLGASGFRKGMDLYFKRHDNKPATCDDFRAAMADANGI 114
+D + A Y KGA V+R+ + LGA F+K + Y K H A +D AA+ +G
Sbjct 364 IDEIFDAISYRKGASVIRMLQSYLGAEVFQKSLAAYIKNHAYSNAKTEDLWAALEAGSGE 423
Query 115 NLDQ-MDRWYSQAGTPRLEVLRSEYKEDEKAFEIS-FRQHTPPTRGQ 159
+++ M W Q G P V+ ++ K+ + E S F P GQ
Sbjct 424 PVNKLMSSWTKQKGYP---VVSAKIKDGKLELEQSRFLSSGSPGEGQ 467
> YHR047c
Length=856
Score = 47.8 bits (112), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 41/154 (26%), Positives = 70/154 (45%), Gaps = 14/154 (9%)
Query 55 MDNFYTATVYDKGAEVVRIYYTLLGASGFRKGMDLYFKRHDNKPATCDDFRAAMADANGI 114
++ + A Y KG+ ++R+ LG F KG+ Y + A D A+ADA+G
Sbjct 377 INQIFDAISYSKGSSLLRMISKWLGEETFIKGVSQYLNKFKYGNAKTGDLWDALADASGK 436
Query 115 NL-DQMDRWYSQAGTPRLEVLRSEYKEDEKAFEISFRQHTPPTRG--QETKRPQLIPIKI 171
++ M+ W + G P L V E+K +I+ QH + G +E + + PI +
Sbjct 437 DVCSVMNIWTKRVGFPVLSV--KEHKN-----KITLTQHRYLSTGDVKEEEDTTIYPILL 489
Query 172 GL---IGRASHQDLLQPAVVLEMTEEEQTFRVRG 202
L G + L + + E+ EE F++ G
Sbjct 490 ALKDSTGIDNTLVLNEKSATFELKNEE-FFKING 522
> Hs20534524
Length=957
Score = 47.8 bits (112), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 39/143 (27%), Positives = 69/143 (48%), Gaps = 17/143 (11%)
Query 1 LKEGLTVFREQLFMGTFMSAGVQRIKE---VADLISRQFPEDDGPMA-HAI-----RPES 51
L EG F E F+G + ++++ + D++ Q EDD M+ H I P+
Sbjct 414 LNEGFASFFE--FLGVNHAETDWQMRDQMLLEDVLPVQ--EDDSLMSSHPIIVTVTTPDE 469
Query 52 YLAMDNFYTATVYDKGAEVVRIYYTLLGASGFRKGMDLYFKRHDNKPATCDDFRAAMADA 111
+ + + Y KG+ ++R+ + F+KG +Y +++ K A DF AA+ +A
Sbjct 470 ---ITSVFDGISYSKGSSILRMLEDWIKPENFQKGCQMYLEKYQFKNAKTSDFWAALEEA 526
Query 112 NGINLDQ-MDRWYSQAGTPRLEV 133
+ + + + MD W Q G P L V
Sbjct 527 SRLPVKEVMDTWTRQMGYPVLNV 549
> Hs4503575
Length=957
Score = 47.4 bits (111), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 39/143 (27%), Positives = 69/143 (48%), Gaps = 17/143 (11%)
Query 1 LKEGLTVFREQLFMGTFMSAGVQRIKE---VADLISRQFPEDDGPMA-HAI-----RPES 51
L EG F E F+G + ++++ + D++ Q EDD M+ H I P+
Sbjct 414 LNEGFASFFE--FLGVNHAETDWQMRDQMLLEDVLPVQ--EDDSLMSSHPIIVTVTTPDE 469
Query 52 YLAMDNFYTATVYDKGAEVVRIYYTLLGASGFRKGMDLYFKRHDNKPATCDDFRAAMADA 111
+ + + Y KG+ ++R+ + F+KG +Y +++ K A DF AA+ +A
Sbjct 470 ---ITSVFDGISYSKGSSILRMLEDWIKPENFQKGCQMYLEKYQFKNAKTSDFWAALEEA 526
Query 112 NGINLDQ-MDRWYSQAGTPRLEV 133
+ + + + MD W Q G P L V
Sbjct 527 SRLPVKEVMDTWTRQMGYPVLNV 549
> YKL157w
Length=935
Score = 47.0 bits (110), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 30/99 (30%), Positives = 49/99 (49%), Gaps = 7/99 (7%)
Query 55 MDNFYTATVYDKGAEVVRIYYTLLGASGFRKGMDLYFKRHDNKPATCDDFRAAMADANGI 114
++ + A Y KGA ++R+ LG F KG+ Y + A +D A+ADA+G
Sbjct 473 INQIFDAISYSKGASLLRMISKWLGEETFIKGVSQYLNKFKYGNAKTEDLWDALADASGK 532
Query 115 NL-DQMDRWYSQAGTPRLEVLRSEYKEDEKAFEISFRQH 152
++ M+ W + G P + V ED +I+FRQ+
Sbjct 533 DVRSVMNIWTKKVGFPVISV-----SEDGNG-KITFRQN 565
> SPBC1921.05
Length=882
Score = 42.4 bits (98), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 30/118 (25%), Positives = 51/118 (43%), Gaps = 3/118 (2%)
Query 55 MDNFYTATVYDKGAEVVRIYYTLLGASGFRKGMDLYFKRHDNKPATCDDFRAAMADANGI 114
++ + A Y KG+ V+R+ +G F KG+ Y +H +D AA++ +G
Sbjct 393 INQIFDAISYSKGSCVIRMVSKYVGEDTFIKGIQKYISKHRYGNTVTEDLWAALSAESGQ 452
Query 115 NLDQ-MDRWYSQAGTPRLEVLRSEYKEDEKAFEISFRQHTPPTRGQETKRPQLIPIKI 171
++ M W + G P L V SE + E E T + +E P+K+
Sbjct 453 DISSTMHNWTKKTGYPVLSV--SETNDGELLIEQHRFLSTGDVKPEEDTVIYWAPLKL 508
> 7299745
Length=913
Score = 41.2 bits (95), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 25/73 (34%), Positives = 35/73 (47%), Gaps = 4/73 (5%)
Query 64 YDKGAEVVRIYYTLLGASGFRKGMDLYFKRHDNKPATCDDFRAAMADANGINLD---QMD 120
Y+KG V+R+ TL+GA F + + Y +H DDF + +A +LD M
Sbjct 451 YEKGGSVIRMLETLVGAEKFEEAVTNYLVKHQFNNTVTDDFLTEV-EAVVTDLDIKKLML 509
Query 121 RWYSQAGTPRLEV 133
W Q G P L V
Sbjct 510 TWTEQMGYPVLNV 522
> 7301772
Length=932
Score = 40.4 bits (93), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 23/81 (28%), Positives = 36/81 (44%), Gaps = 9/81 (11%)
Query 64 YDKGAEVVRIYYTLLGASGFRKGMDLYFKRHDNKPATCDDFRAAMADA--------NGIN 115
Y KG+ V+R+ + LG FR G+ Y ++ K A D+ ++ A +
Sbjct 428 YQKGSTVLRMMHLFLGEESFRSGLQAYLQKFSYKNAEQDNLWESLTQAAHKYRSLPKSYD 487
Query 116 LDQ-MDRWYSQAGTPRLEVLR 135
+ MD W Q G P + V R
Sbjct 488 IKSIMDSWTLQTGYPVINVTR 508
> CE21162
Length=988
Score = 40.4 bits (93), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 41/152 (26%), Positives = 61/152 (40%), Gaps = 20/152 (13%)
Query 1 LKEGLTVFREQLFMGT-FMSAGVQRIKE--VADLISRQFPEDDGPMAHAIRPESYLAM-- 55
L EG E ++GT +S G R++E D + D H + + AM
Sbjct 417 LNEGFATLVE--YLGTDEISDGNMRMREWFTMDALWSALAADSVASTHPLTFKIDKAMEV 474
Query 56 -DNFYTATVYDKGAEVVRIYYTLLGASGFRKGMDLYFKRHDNKPATCDDFRAAMADA--- 111
D+F + T YDKG V+ + +G F G++ Y RH A + A+ +
Sbjct 475 LDSFDSVT-YDKGGAVLAMVRKTIGEENFNTGINHYLTRHQFDNADAGNLLTALGEKIPD 533
Query 112 -----NGINLDQ---MDRWYSQAGTPRLEVLR 135
G+ L+ MD W Q G P L R
Sbjct 534 SVMGPKGVKLNISEFMDPWTKQLGYPLLNASR 565
> 7301771
Length=1082
Score = 40.0 bits (92), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 27/87 (31%), Positives = 40/87 (45%), Gaps = 11/87 (12%)
Query 59 YTATVYDKGAEVVRIYYTLLGASGFRKGMDLYFKRHDNKPATCDDFRAAMADA---NG-- 113
+ A Y KG+ V+R+ + +G FR G+ Y K + K A ++ ++ A NG
Sbjct 418 FDAISYQKGSAVLRMMHLFMGEESFRTGLREYLKLYAYKNAEQNNLWESLTTAAHQNGAL 477
Query 114 -----INLDQMDRWYSQAGTPRLEVLR 135
IN MD W Q G P L + R
Sbjct 478 PGHYDIN-TIMDSWTLQTGFPVLNITR 503
> Hs22064092
Length=478
Score = 40.0 bits (92), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 26/93 (27%), Positives = 44/93 (47%), Gaps = 8/93 (8%)
Query 7 VFREQLFMGTFMSAGVQRIKEVADLISRQFPEDDGPMAHAIRPESYLAMDNFYTATVYDK 66
F E F+SA R +E+ L ++ P+ ++ S +D + A Y K
Sbjct 389 CFPEYDIWTQFVSADYTRAQELDAL------DNSHPIEVSVGHPS--EVDEIFDAISYSK 440
Query 67 GAEVVRIYYTLLGASGFRKGMDLYFKRHDNKPA 99
GA V+R+ + +G F+KGM++Y + K A
Sbjct 441 GASVIRMLHDYIGDKDFKKGMNMYLTKFQQKNA 473
> 7301751
Length=1017
Score = 39.3 bits (90), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 37/146 (25%), Positives = 57/146 (39%), Gaps = 11/146 (7%)
Query 1 LKEGLTVFREQLFMGTFMSAGVQRIKEVADLISRQFPEDDGPMAHAIRPESY--LAMDNF 58
L EG + E L Q + V + + F D +H I E + +
Sbjct 455 LNEGFASYMEYLTADAVAPEWKQLDQFVVNELQAVFQLDALSTSHKISHEVFNPQEISEI 514
Query 59 YTATVYDKGAEVVRIYYTLLGASGFRKGMDLYFKRHDNKPATCDD---FRAAMADANGIN 115
+ Y KG+ ++R+ L FR+G+ Y + AT DD F A ++G+
Sbjct 515 FDRISYAKGSTIIRMMAHFLTNPIFRRGLSKYLQEMAYNSATQDDLWHFLTVEAKSSGLL 574
Query 116 LDQ------MDRWYSQAGTPRLEVLR 135
D MD W Q G P ++V R
Sbjct 575 DDSRSVKEIMDTWTLQTGYPVVKVSR 600
> ECU01g1470
Length=864
Score = 38.9 bits (89), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 23/80 (28%), Positives = 37/80 (46%), Gaps = 1/80 (1%)
Query 55 MDNFYTATVYDKGAEVVRIYYTLLGASGFRKGMDLYFKRHDNKPATCDDFRAAMADANGI 114
+ + + Y KGA V+R+ +G S F G+ Y K H A+ + G
Sbjct 404 IGEIFDSISYCKGASVIRMIERYVGESVFMLGIRRYIKEHMYGNGNAMSLWKAIGEEYGE 463
Query 115 NLDQM-DRWYSQAGTPRLEV 133
++ +M + W SQAG P + V
Sbjct 464 DISEMVEGWISQAGYPVVSV 483
> ECU01g0140
Length=864
Score = 38.9 bits (89), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 23/80 (28%), Positives = 37/80 (46%), Gaps = 1/80 (1%)
Query 55 MDNFYTATVYDKGAEVVRIYYTLLGASGFRKGMDLYFKRHDNKPATCDDFRAAMADANGI 114
+ + + Y KGA V+R+ +G S F G+ Y K H A+ + G
Sbjct 404 IGEIFDSISYCKGASVIRMIERYVGESVFMLGIRRYIKEHMYGNGNAMSLWKAIGEEYGE 463
Query 115 NLDQM-DRWYSQAGTPRLEV 133
++ +M + W SQAG P + V
Sbjct 464 DISEMVEGWISQAGYPVVSV 483
> ECU08g0070
Length=864
Score = 38.9 bits (89), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 23/80 (28%), Positives = 37/80 (46%), Gaps = 1/80 (1%)
Query 55 MDNFYTATVYDKGAEVVRIYYTLLGASGFRKGMDLYFKRHDNKPATCDDFRAAMADANGI 114
+ + + Y KGA V+R+ +G S F G+ Y K H A+ + G
Sbjct 404 IGEIFDSISYCKGASVIRMIERYVGESVFMLGIRRYIKEHMYGNGNAMSLWKAIGEEYGE 463
Query 115 NLDQM-DRWYSQAGTPRLEV 133
++ +M + W SQAG P + V
Sbjct 464 DISEMVEGWISQAGYPVVSV 483
> 7291558
Length=844
Score = 37.4 bits (85), Expect = 0.035, Method: Compositional matrix adjust.
Identities = 28/105 (26%), Positives = 48/105 (45%), Gaps = 12/105 (11%)
Query 59 YTATVYDKGAEVVRIYYTLLGASGFRKGMDLYFKRHDNKPATCDDFRAAMADA------- 111
+T VY+KG+ +R+ + L+G F G+ + +R D ++ A
Sbjct 349 FTEYVYEKGSLTIRMLHKLVGEEAFFHGIRSFLERFSFGNVAQADLWNSLQMAALKNQVI 408
Query 112 -NGINLDQ-MDRWYSQAGTPRLEVLRSEYKEDEKAFEIS--FRQH 152
+ NL + MD W Q G P + ++R+ YK E S F++H
Sbjct 409 SSDFNLSRAMDSWTLQGGYPLVTLIRN-YKTGEVTLNQSRFFQEH 452
> 7300768
Length=1878
Score = 35.0 bits (79), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 26/103 (25%), Positives = 43/103 (41%), Gaps = 10/103 (9%)
Query 42 PMAHAIRPESYLAMDNFYTATVYDKGAEVVRIYYTLLGASGFRKGMDLYFKRHDNKPATC 101
PM+H ++ S +A+ Y + Y K V+ ++ L + F++G+ Y + A
Sbjct 1347 PMSHFVQKPSEIAL--LYDSVSYAKAGSVLDMWRHALTNTVFQRGLHNYLVDNQFTAANP 1404
Query 102 DDFRAAMADA--------NGINLDQMDRWYSQAGTPRLEVLRS 136
A+A A D M W +Q G P L V R+
Sbjct 1405 SKLFEAIAKAAVEENYEVKATVPDMMGSWTNQGGVPLLTVTRN 1447
> 7299744
Length=988
Score = 34.7 bits (78), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 24/87 (27%), Positives = 40/87 (45%), Gaps = 5/87 (5%)
Query 64 YDKGAEVVRIYYTLLGASGFRKGMDLYFKRHDNKPATCDDFRAAMADAN-GINLDQ-MDR 121
Y KG+ +VR+ LG + FR+ + Y + A +F + G N+ + M
Sbjct 506 YSKGSSLVRMLEDFLGETTFRQAVTNYLNEYKYSTAETGNFFTEIDKLELGYNVTEIMLT 565
Query 122 WYSQAGTPRLEVLR---SEYKEDEKAF 145
W Q G P + + + +EYK +K F
Sbjct 566 WTVQMGLPVVTIEKVSDTEYKLTQKRF 592
> CE27507
Length=1073
Score = 34.7 bits (78), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 31/145 (21%), Positives = 62/145 (42%), Gaps = 10/145 (6%)
Query 1 LKEGLTVFREQLFMGTF--MSAGVQRIKEVADLISRQFPEDDG-PMAHAIRPESYLAMDN 57
L EG F M T ++A + +A LI Q ED + + ES + +
Sbjct 445 LNEGFATFYVYEMMSTERPVTAQFEYYDSLASLILAQSEEDHRLSLVRELATESQVE-SS 503
Query 58 FYTATVYDKGAEVVRIYYTLLGASGFRKGMDLYFKRHDNKPATCDDFRAAMADANGINLD 117
F+ +Y KG ++R+ L+ F+ + Y +++ + + DD A++ +
Sbjct 504 FHPTNLYTKGCVLIRMLRDLVSDFDFKAAVRRYLRKNAYRSVSRDDLFASLPAYADHGAE 563
Query 118 Q------MDRWYSQAGTPRLEVLRS 136
Q ++ W+ G P + ++R+
Sbjct 564 QEKLSHVLEGWFVNEGIPEITLIRN 588
> CE18905
Length=747
Score = 34.3 bits (77), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 29/106 (27%), Positives = 47/106 (44%), Gaps = 15/106 (14%)
Query 39 DDGPMAHAIRPESYLAMDNFYTATVYDKGAEVVRIYYTLLGASGFRKGMDLYFKRHDNKP 98
+DG ++ M+N ATVY KG VR+ + G F + + Y ++ N+
Sbjct 385 NDGGVSLNFGKNLSSVMNN--AATVYMKGCSFVRMLENIFGTEYFNEAIKFYLEK--NQY 440
Query 99 ATCDD------FRAA--MADANG--INLDQMDR-WYSQAGTPRLEV 133
DD FR + D NG ++++ R W Q G P ++V
Sbjct 441 DNVDDNDLYEAFRTSSETLDTNGKPFDIEKFARCWTHQNGFPTIQV 486
> Hs20149637
Length=948
Score = 33.9 bits (76), Expect = 0.37, Method: Compositional matrix adjust.
Identities = 15/46 (32%), Positives = 24/46 (52%), Gaps = 0/46 (0%)
Query 64 YDKGAEVVRIYYTLLGASGFRKGMDLYFKRHDNKPATCDDFRAAMA 109
YDKGA ++ + L A F+ G+ Y ++H K +D +MA
Sbjct 438 YDKGACILNMLREYLSADAFKSGIVQYLQKHSYKNTKNEDLWDSMA 483
> Hs11641261
Length=960
Score = 33.9 bits (76), Expect = 0.39, Method: Compositional matrix adjust.
Identities = 14/48 (29%), Positives = 28/48 (58%), Gaps = 0/48 (0%)
Query 64 YDKGAEVVRIYYTLLGASGFRKGMDLYFKRHDNKPATCDDFRAAMADA 111
Y+KGA ++ + LG F+KG+ Y K+ + A DD ++++++
Sbjct 455 YNKGACILNMLKDFLGEEKFQKGIIQYLKKFSYRNAKNDDLWSSLSNS 502
> 7294697
Length=887
Score = 33.5 bits (75), Expect = 0.58, Method: Compositional matrix adjust.
Identities = 23/84 (27%), Positives = 37/84 (44%), Gaps = 9/84 (10%)
Query 66 KGAEVVRIYYTLLGASGFRKGMDLYFKRHDNKPATCDDF--------RAAMADANGINLD 117
K ++ + LG F KG+ +FKR+ N A+ + R + G++L
Sbjct 405 KMCLLIHMLKVALGDKLFLKGLQEFFKRYANSSASSKELWEEFQRGARRSHQLPTGVSLP 464
Query 118 Q-MDRWYSQAGTPRLEVLRSEYKE 140
M+ W Q G P L V R + K+
Sbjct 465 TVMESWMKQPGFPLLTVQRDDAKQ 488
> 7298498
Length=613
Score = 33.1 bits (74), Expect = 0.66, Method: Compositional matrix adjust.
Identities = 36/147 (24%), Positives = 58/147 (39%), Gaps = 23/147 (15%)
Query 1 LKEGLTVFREQLFMGTFMSAGVQRIKEVADLISRQ---------FPEDDGPMAHAIRPES 51
L EG TVF E +G A K +++L Q PE + + S
Sbjct 314 LNEGFTVFVESKIVGRMQGAKELDFKMLSNLTDLQECIRTQLNKTPE----LTKLVVDLS 369
Query 52 YLAMDNFYTATVYDKGAEVVRIYYTLLGA-SGFRKGMDLYFKRHDNKPATCDDFRAAMAD 110
D+ +++ Y KG+ +R L G + F + Y K++ K DF++A+ D
Sbjct 370 NCGPDDAFSSVPYIKGSTFLRYLEDLFGGPTVFEPFLRDYLKKYAYKSIETKDFQSALYD 429
Query 111 ANGINLDQMDR--------WYSQAGTP 129
I+ D+ D+ W G P
Sbjct 430 Y-FIDTDKKDKLSAVDWDLWLKSEGMP 455
> Hs5031881
Length=1025
Score = 32.7 bits (73), Expect = 0.99, Method: Compositional matrix adjust.
Identities = 46/207 (22%), Positives = 78/207 (37%), Gaps = 18/207 (8%)
Query 1 LKEGLTVFREQLFMGTFMSAGVQRIKEVADLISRQFP---EDDGPMAHAIRP--ESYLAM 55
L EG F E + + + D + +F +D +H I +S +
Sbjct 485 LNEGFATFMEYFSLEKIF----KELSSYEDFLDARFKTMKKDSLNSSHPISSSVQSSEQI 540
Query 56 DNFYTATVYDKGAEVVRIYYTLLGASGFRKGMDLYFKRHDNKPATCDDFRAAMADANGIN 115
+ + + Y KG+ ++ + T L F+ + LY H DD + +
Sbjct 541 EEMFDSLSYFKGSSLLLMLKTYLSEDVFQHAVVLYLHNHSYASIQSDDLWDSFNEVTNQT 600
Query 116 LD---QMDRWYSQAGTPRLEVLRSEYKEDEKAFEISFRQHTPPTRGQETKRPQLIPIKIG 172
LD M W Q G P + V + + KE E F P + +T IP+
Sbjct 601 LDVKRMMKTWTLQKGFPLVTV-QKKGKELFIQQERFFLNMKPEIQPSDTSYLWHIPLSYV 659
Query 173 LIGR--ASHQD---LLQPAVVLEMTEE 194
GR + +Q L + + V+ +TEE
Sbjct 660 TEGRNYSKYQSVSLLDKKSGVINLTEE 686
> CE20255
Length=1045
Score = 31.6 bits (70), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 25/89 (28%), Positives = 36/89 (40%), Gaps = 12/89 (13%)
Query 61 ATVYDKGAEVVRIYYTLLGASGFRKGMDLYFKRHDNKPATCDDF---------RAAMADA 111
A +Y K A ++R+ L+ FR+G+ + K A +D +A
Sbjct 543 AIIYKKSAIIIRMIERLVEEDIFRQGLKRFLNTFLYKNADHEDLFNVLVYVHDSSAGGHL 602
Query 112 NGINL---DQMDRWYSQAGTPRLEVLRSE 137
G N D MD W QA P + V R E
Sbjct 603 RGQNFSLSDVMDTWIRQAHFPVIHVNRKE 631
> Hs22040930
Length=168
Score = 30.8 bits (68), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 21/51 (41%), Positives = 24/51 (47%), Gaps = 7/51 (13%)
Query 147 ISFRQHTPPTRGQETKRPQLIPIKIGLIGRASHQDLLQPA---VVLEMTEE 194
IS R H PT G + RP L P G SH+ L P V +E TEE
Sbjct 94 ISHRPHLSPTPGPLSHRPHLSPTP----GPISHRPHLSPTAEYVCVEFTEE 140
> CE26001
Length=1815
Score = 30.4 bits (67), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 14/49 (28%), Positives = 26/49 (53%), Gaps = 0/49 (0%)
Query 63 VYDKGAEVVRIYYTLLGASGFRKGMDLYFKRHDNKPATCDDFRAAMADA 111
Y+KGA ++R+ +LGA F++G+ Y ++ A D + + D
Sbjct 471 TYNKGASMLRMLSDVLGADVFKQGIRAYLQKMQYSNANDFDLFSTLTDT 519
> CE29474
Length=700
Score = 30.0 bits (66), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 12/36 (33%), Positives = 26/36 (72%), Gaps = 2/36 (5%)
Query 165 QLIPIKIGLIG--RASHQDLLQPAVVLEMTEEEQTF 198
++IP+K+G+IG S Q++L ++++ ++EQ+F
Sbjct 235 KVIPVKLGIIGVVNRSQQNILDNKLIVDAVKDEQSF 270
> At2g17290
Length=544
Score = 29.6 bits (65), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 19/62 (30%), Positives = 29/62 (46%), Gaps = 11/62 (17%)
Query 245 WFASRALA-----TPIVISRSKRISAKGNAQKL------EQLPDDYVEGLRAIIYDQATD 293
W +A P V+SR K+ SA +K+ E L ++ + GLRA+ TD
Sbjct 342 WICENGVAPDRALDPAVLSRLKQFSAMNKLKKMALKVIAESLSEEEIAGLRAMFEAMDTD 401
Query 294 NA 295
N+
Sbjct 402 NS 403
> Hs17449263
Length=144
Score = 29.3 bits (64), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 24/74 (32%), Positives = 35/74 (47%), Gaps = 17/74 (22%)
Query 204 REDCVPSILRGFSAPVRLINRPQTAKESAFLLAYDTDPV-TKWFASRALATPIVISRSKR 262
R+ P IL G S +T +ES PV + AS+ +TP ISR+
Sbjct 47 RKTLTPKILEGAS---------KTGRES-------RQPVRPQACASKERSTPSTISRAAE 90
Query 263 ISAKGNAQKLEQLP 276
+SA GN +++E P
Sbjct 91 LSASGNEREIEDFP 104
Lambda K H
0.320 0.135 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6602058562
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40