bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0510_orf1
Length=131
Score E
Sequences producing significant alignments: (Bits) Value
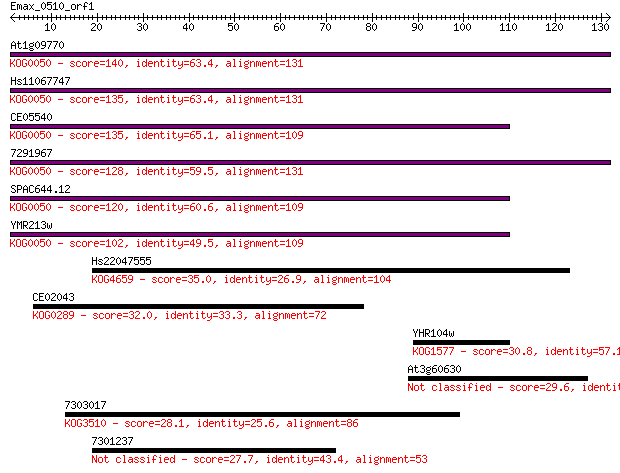
At1g09770 140 5e-34
Hs11067747 135 1e-32
CE05540 135 2e-32
7291967 128 2e-30
SPAC644.12 120 6e-28
YMR213w 102 1e-22
Hs22047555 35.0 0.035
CE02043 32.0 0.29
YHR104w 30.8 0.78
At3g60630 29.6 1.5
7303017 28.1 4.4
7301237 27.7 6.6
> At1g09770
Length=844
Score = 140 bits (354), Expect = 5e-34, Method: Compositional matrix adjust.
Identities = 83/131 (63%), Positives = 100/131 (76%), Gaps = 1/131 (0%)
Query 1 LRPGEIDPHPETKPSRADPIDMAEDEKEMLEEARARLANTRGKKAKRKAREKQLEEARRL 60
LRPGEIDP+PE KP+R DP+DM EDEKEML EARARLANTRGKKAKRKAREKQLEEARRL
Sbjct 122 LRPGEIDPNPEAKPARPDPVDMDEDEKEMLSEARARLANTRGKKAKRKAREKQLEEARRL 181
Query 61 AALQKKRELKAAGIITGAKRRTRKNFQPYEEVPFEEKPPPGFYEVPSEENPEGNLNFANI 120
A+LQK+RELKAAGI ++R RK E+PFE++ P GFY+ E+ P + F
Sbjct 182 ASLQKRRELKAAGIDGRHRKRKRKGIDYNAEIPFEKRAPAGFYDTADEDRPADQVKFPT- 240
Query 121 SLQHMEGHMRA 131
+++ +EG RA
Sbjct 241 TIEELEGKRRA 251
> Hs11067747
Length=802
Score = 135 bits (341), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 83/132 (62%), Positives = 103/132 (78%), Gaps = 2/132 (1%)
Query 1 LRPGEIDPHPETKPSRADPIDMAEDEKEMLEEARARLANTRGKKAKRKAREKQLEEARRL 60
L+PGEIDP+PETKP+R DPIDM EDE EML EARARLANT+GKKAKRKAREKQLEEARRL
Sbjct 123 LKPGEIDPNPETKPARPDPIDMDEDELEMLSEARARLANTQGKKAKRKAREKQLEEARRL 182
Query 61 AALQKKRELKAAGIITGAKRRTRKNFQPYEEVPFEEKPPPGFYEVPSEENPEG-NLNFAN 119
AALQK+REL+AAGI KR+ ++ E+PFE+KP GFY+ SEEN + + +F
Sbjct 183 AALQKRRELRAAGIEIQKKRKRKRGVDYNAEIPFEKKPALGFYDT-SEENYQALDADFRK 241
Query 120 ISLQHMEGHMRA 131
+ Q ++G +R+
Sbjct 242 LRQQDLDGELRS 253
> CE05540
Length=755
Score = 135 bits (339), Expect = 2e-32, Method: Compositional matrix adjust.
Identities = 71/110 (64%), Positives = 89/110 (80%), Gaps = 2/110 (1%)
Query 1 LRPGEIDPHPETKPSRADPIDMAEDEKEMLEEARARLANTRGKKAKRKAREKQLEEARRL 60
L+PGEIDP PETKP+R DPIDM +DE EML EARARLANT+GKKAKRKARE+QL +ARRL
Sbjct 124 LKPGEIDPTPETKPARPDPIDMDDDELEMLSEARARLANTQGKKAKRKARERQLSDARRL 183
Query 61 AALQKKRELKAAGIITGAKRRTRKNFQPY-EEVPFEEKPPPGFYEVPSEE 109
A+LQK+RE++AAG+ K + ++N Y EE+PFE+ P GF+ PSE+
Sbjct 184 ASLQKRREMRAAGLAFARKFKPKRNQIDYSEEIPFEKHVPAGFHN-PSED 232
> 7291967
Length=791
Score = 128 bits (322), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 78/131 (59%), Positives = 98/131 (74%), Gaps = 1/131 (0%)
Query 1 LRPGEIDPHPETKPSRADPIDMAEDEKEMLEEARARLANTRGKKAKRKAREKQLEEARRL 60
L+PGEIDP+PETKP+R DP DM EDE EML EARARLANT+GKKAKRKAREKQLEEARRL
Sbjct 100 LKPGEIDPNPETKPARPDPKDMDEDELEMLSEARARLANTQGKKAKRKAREKQLEEARRL 159
Query 61 AALQKKRELKAAGIITGAKRRTRKNFQPYEEVPFEEKPPPGFYEVPSEENPEGNLNFANI 120
A LQK+REL+AAGI +G ++R K E+PFE++P GFY+ E + +F +
Sbjct 160 ATLQKRRELRAAGIGSGNRKRI-KGIDYNAEIPFEKRPAHGFYDTSEEHLQKNEPDFNKM 218
Query 121 SLQHMEGHMRA 131
Q ++G +R+
Sbjct 219 RQQDLDGELRS 229
> SPAC644.12
Length=757
Score = 120 bits (301), Expect = 6e-28, Method: Compositional matrix adjust.
Identities = 66/109 (60%), Positives = 77/109 (70%), Gaps = 0/109 (0%)
Query 1 LRPGEIDPHPETKPSRADPIDMAEDEKEMLEEARARLANTRGKKAKRKAREKQLEEARRL 60
LR GE +P+ ET P+ D IDM EDEKEML EARARLANT+GKKAKRK REKQLE RRL
Sbjct 134 LRFGEAEPNLETLPALPDAIDMDEDEKEMLSEARARLANTQGKKAKRKDREKQLELTRRL 193
Query 61 AALQKKRELKAAGIITGAKRRTRKNFQPYEEVPFEEKPPPGFYEVPSEE 109
+ LQK+RELKAAGI RR + +PFE+KP GFY+ E+
Sbjct 194 SHLQKRRELKAAGINIKLFRRKKNEMDYNASIPFEKKPAIGFYDTSEED 242
> YMR213w
Length=590
Score = 102 bits (255), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 54/112 (48%), Positives = 79/112 (70%), Gaps = 3/112 (2%)
Query 1 LRPGEIDPHPETKPSRADPIDMAEDEKEMLEEARARLANTRGKKAKRKAREKQLEEARRL 60
L+ G+I+P+ ET+ +R D D+ ++EKEML EARARL NT+GKKA RK RE+ LEE++R+
Sbjct 123 LKAGDINPNAETQMARPDNGDLEDEEKEMLAEARARLLNTQGKKATRKIRERMLEESKRI 182
Query 61 AALQKKRELKAAGIITGAKRRTRK---NFQPYEEVPFEEKPPPGFYEVPSEE 109
A LQK+RELK AGI K+ +K + E++ +E+ P PG Y+ +E+
Sbjct 183 AELQKRRELKQAGINVAIKKPKKKYGTDIDYNEDIVYEQAPMPGIYDTSTED 234
> Hs22047555
Length=1787
Score = 35.0 bits (79), Expect = 0.035, Method: Composition-based stats.
Identities = 28/104 (26%), Positives = 48/104 (46%), Gaps = 23/104 (22%)
Query 19 PIDMAEDEKEMLEEARARLANTRGKKAKRKAREKQLEEARRLAALQKKRELKAAGIITGA 78
P + E++ +L++AR R T K ++KAR+ + E RL +K++L + G + G
Sbjct 1698 PDTLDEEKARVLDQARQRALGTAWAKEQQKARDGR--EGSRLWTEGEKQQLLSTGRVQG- 1754
Query 79 KRRTRKNFQPYEEVPFEEKPPPGFYEVPSEENPEGNLNFANISL 122
YE G+Y +P E+ PE + +NI
Sbjct 1755 ----------YE----------GYYVLPVEQYPELADSSSNIQF 1778
> CE02043
Length=509
Score = 32.0 bits (71), Expect = 0.29, Method: Composition-based stats.
Identities = 24/73 (32%), Positives = 35/73 (47%), Gaps = 3/73 (4%)
Query 6 IDPHPETKPSRADPIDMAEDEKEMLEEARARL-ANTRGKKAKRKAREKQLEEARRLAALQ 64
+ PH K ID +ED++ + E A+L ++ A+RK R K L E LA +
Sbjct 152 LKPHTSAKVDDDVSIDESEDQQGLSEAILAKLEEKSKSLTAERKQRGKNLPEG--LAKTE 209
Query 65 KKRELKAAGIITG 77
+ ELK TG
Sbjct 210 ELAELKQTASHTG 222
> YHR104w
Length=327
Score = 30.8 bits (68), Expect = 0.78, Method: Compositional matrix adjust.
Identities = 12/21 (57%), Positives = 15/21 (71%), Gaps = 0/21 (0%)
Query 89 YEEVPFEEKPPPGFYEVPSEE 109
++ VPFEEK PPGFY +E
Sbjct 116 FKYVPFEEKYPPGFYTGADDE 136
> At3g60630
Length=623
Score = 29.6 bits (65), Expect = 1.5, Method: Composition-based stats.
Identities = 15/45 (33%), Positives = 20/45 (44%), Gaps = 6/45 (13%)
Query 88 PYEEVPFEEK------PPPGFYEVPSEENPEGNLNFANISLQHME 126
P++ P EEK P PGF+ P P LN QH++
Sbjct 163 PFQNAPEEEKFQISINPNPGFFSDPPSSPPAKRLNSGQPGSQHLQ 207
> 7303017
Length=2542
Score = 28.1 bits (61), Expect = 4.4, Method: Composition-based stats.
Identities = 22/86 (25%), Positives = 41/86 (47%), Gaps = 6/86 (6%)
Query 13 KPSRADPIDMAEDEKEMLEEARARLANTRGKKAKRKAREKQLEEARRLAALQKKRELKAA 72
KP ++ + E+ + E + + +T K K A +KQ+ E + L A +K+
Sbjct 2404 KPEVSEVVAEKISEETIEEPKKPEVKDTEIKSEKATALDKQVLEEKELEASAQKQ----- 2458
Query 73 GIITGAKRRTRKNFQPYEEVPFEEKP 98
+++++K +PYE P EKP
Sbjct 2459 -CDQDVEKKSQKPERPYENDPKWEKP 2483
> 7301237
Length=572
Score = 27.7 bits (60), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 23/68 (33%), Positives = 37/68 (54%), Gaps = 17/68 (25%)
Query 19 PIDMA---EDEKEML------------EEARARLANTRGKKAKRKAREKQLEEARRLAAL 63
P+DM EDEK++ +EA+ A+T G+K + K ++Q +EA + AAL
Sbjct 254 PLDMDSQEEDEKDIQGGEAKTNAEVFEDEAQTSAADTDGQKHEEK--DEQADEATKPAAL 311
Query 64 QKKRELKA 71
K +E+ A
Sbjct 312 DKDKEVTA 319
Lambda K H
0.311 0.130 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1283105810
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40