bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0420_orf1
Length=145
Score E
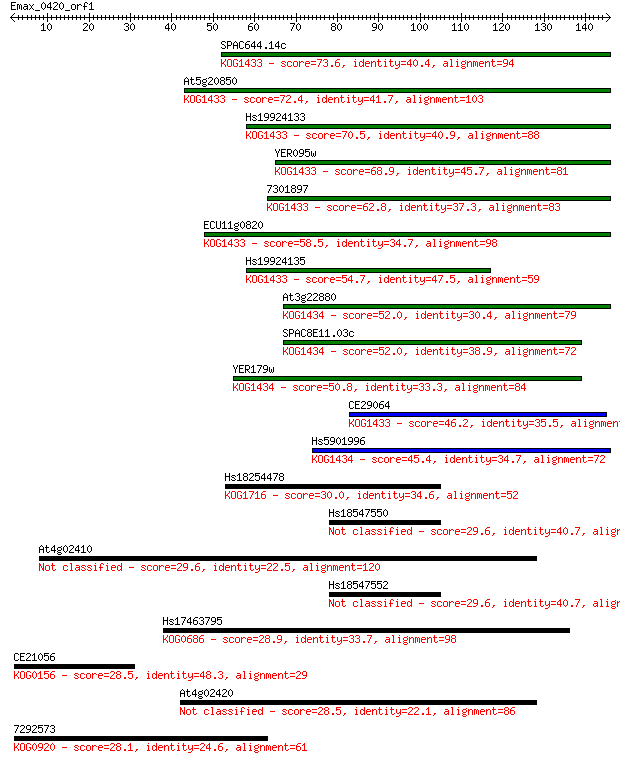
Sequences producing significant alignments: (Bits) Value
SPAC644.14c 73.6 1e-13
At5g20850 72.4 3e-13
Hs19924133 70.5 1e-12
YER095w 68.9 3e-12
7301897 62.8 2e-10
ECU11g0820 58.5 4e-09
Hs19924135 54.7 6e-08
At3g22880 52.0 4e-07
SPAC8E11.03c 52.0 4e-07
YER179w 50.8 8e-07
CE29064 46.2 2e-05
Hs5901996 45.4 3e-05
Hs18254478 30.0 1.6
Hs18547550 29.6 1.8
At4g02410 29.6 1.9
Hs18547552 29.6 1.9
Hs17463795 28.9 3.7
CE21056 28.5 4.1
At4g02420 28.5 4.9
7292573 28.1 5.7
> SPAC644.14c
Length=365
Score = 73.6 bits (179), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 38/94 (40%), Positives = 55/94 (58%), Gaps = 0/94 (0%)
Query 52 QLQDVGEDLQTGPLKLEHLLAKGLTRKDLDLLKEGGLHTVECVAFAPLKALLAIKGISEQ 111
+QD ++ GP+ L+ L G+T D+ + E G +TVE +A+ P + LL IKGISE
Sbjct 32 NVQDEEDEAAAGPMPLQMLEGNGITASDIKKIHEAGYYTVESIAYTPKRQLLLIKGISEA 91
Query 112 KAAKLKQASKELCSLGFCSAQEYLEARANLIKFT 145
KA KL + +L +GF +A EY R+ LI T
Sbjct 92 KADKLLGEASKLVPMGFTTATEYHIRRSELITIT 125
> At5g20850
Length=339
Score = 72.4 bits (176), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 43/103 (41%), Positives = 58/103 (56%), Gaps = 2/103 (1%)
Query 43 QETRQAPEQQLQDVGEDLQTGPLKLEHLLAKGLTRKDLDLLKEGGLHTVECVAFAPLKAL 102
Q Q QQ D E+ Q GP +E L A G+ D+ L++ GL TVE VA+ P K L
Sbjct 3 QRRNQNAVQQQDD--EETQHGPFPVEQLQAAGIASVDVKKLRDAGLCTVEGVAYTPRKDL 60
Query 103 LAIKGISEQKAAKLKQASKELCSLGFCSAQEYLEARANLIKFT 145
L IKGIS+ K K+ +A+ +L LGF SA + R +I+ T
Sbjct 61 LQIKGISDAKVDKIVEAASKLVPLGFTSASQLHAQRQEIIQIT 103
> Hs19924133
Length=339
Score = 70.5 bits (171), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 36/88 (40%), Positives = 53/88 (60%), Gaps = 0/88 (0%)
Query 58 EDLQTGPLKLEHLLAKGLTRKDLDLLKEGGLHTVECVAFAPLKALLAIKGISEQKAAKLK 117
E+ GP + L G+ D+ L+E G HTVE VA+AP K L+ IKGISE KA K+
Sbjct 16 EEESFGPQPISRLEQCGINANDVKKLEEAGFHTVEAVAYAPKKELINIKGISEAKADKIL 75
Query 118 QASKELCSLGFCSAQEYLEARANLIKFT 145
+ +L +GF +A E+ + R+ +I+ T
Sbjct 76 AEAAKLVPMGFTTATEFHQRRSEIIQIT 103
> YER095w
Length=400
Score = 68.9 bits (167), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 37/81 (45%), Positives = 49/81 (60%), Gaps = 0/81 (0%)
Query 65 LKLEHLLAKGLTRKDLDLLKEGGLHTVECVAFAPLKALLAIKGISEQKAAKLKQASKELC 124
+ +E L G+T D+ L+E GLHT E VA+AP K LL IKGISE KA KL + L
Sbjct 81 VPIEKLQVNGITMADVKKLRESGLHTAEAVAYAPRKDLLEIKGISEAKADKLLNEAARLV 140
Query 125 SLGFCSAQEYLEARANLIKFT 145
+GF +A ++ R+ LI T
Sbjct 141 PMGFVTAADFHMRRSELICLT 161
> 7301897
Length=336
Score = 62.8 bits (151), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 31/83 (37%), Positives = 51/83 (61%), Gaps = 0/83 (0%)
Query 63 GPLKLEHLLAKGLTRKDLDLLKEGGLHTVECVAFAPLKALLAIKGISEQKAAKLKQASKE 122
GPL + L+ +T KD+ LL++ LHTVE VA A K L+AI G+ K ++ + +
Sbjct 18 GPLSVTKLIGGSITAKDIKLLQQASLHTVESVANATKKQLMAIPGLGGGKVEQIITEANK 77
Query 123 LCSLGFCSAQEYLEARANLIKFT 145
L LGF SA+ + + RA++++ +
Sbjct 78 LVPLGFLSARTFYQMRADVVQLS 100
> ECU11g0820
Length=334
Score = 58.5 bits (140), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 34/98 (34%), Positives = 53/98 (54%), Gaps = 2/98 (2%)
Query 48 APEQQLQDVGEDLQTGPLKLEHLLAKGLTRKDLDLLKEGGLHTVECVAFAPLKALLAIKG 107
+ E Q QD + + + + L G+ D+ L E G TVE +AFAP + LL+IKG
Sbjct 2 SEEYQYQDT--EYHSESISISELKNGGILAVDIAKLIEAGFTTVESLAFAPKRQLLSIKG 59
Query 108 ISEQKAAKLKQASKELCSLGFCSAQEYLEARANLIKFT 145
S+ K KL + + +L +GF +A Y + R+ L+ T
Sbjct 60 FSDIKVDKLIKEAAKLVPMGFTTASAYHQRRSELVYLT 97
> Hs19924135
Length=242
Score = 54.7 bits (130), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 28/59 (47%), Positives = 36/59 (61%), Gaps = 0/59 (0%)
Query 58 EDLQTGPLKLEHLLAKGLTRKDLDLLKEGGLHTVECVAFAPLKALLAIKGISEQKAAKL 116
E+ GP + L G+ D+ L+E G HTVE VA+AP K L+ IKGISE KA K+
Sbjct 16 EEESFGPQPISRLEQCGINANDVKKLEEAGFHTVEAVAYAPKKELINIKGISEAKADKI 74
> At3g22880
Length=339
Score = 52.0 bits (123), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 24/79 (30%), Positives = 47/79 (59%), Gaps = 0/79 (0%)
Query 67 LEHLLAKGLTRKDLDLLKEGGLHTVECVAFAPLKALLAIKGISEQKAAKLKQASKELCSL 126
++ L+A+G+ D+ L+E G+HT + K L IKG+SE K K+ +A++++ +
Sbjct 31 IDKLIAQGINAGDVKKLQEAGIHTCNGLMMHTKKNLTGIKGLSEAKVDKICEAAEKIVNF 90
Query 127 GFCSAQEYLEARANLIKFT 145
G+ + + L R +++K T
Sbjct 91 GYMTGSDALIKRKSVVKIT 109
> SPAC8E11.03c
Length=332
Score = 52.0 bits (123), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 28/72 (38%), Positives = 42/72 (58%), Gaps = 0/72 (0%)
Query 67 LEHLLAKGLTRKDLDLLKEGGLHTVECVAFAPLKALLAIKGISEQKAAKLKQASKELCSL 126
+E L A G+ D+ LK+ G+ TV+ V + + LL IKG SE K KLK+A+ ++C
Sbjct 18 IEDLTAHGIGMTDIIKLKQAGVCTVQGVHMSTKRFLLKIKGFSEAKVDKLKEAASKMCPA 77
Query 127 GFCSAQEYLEAR 138
F +A E + R
Sbjct 78 NFSTAMEISQNR 89
> YER179w
Length=334
Score = 50.8 bits (120), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 28/84 (33%), Positives = 44/84 (52%), Gaps = 0/84 (0%)
Query 55 DVGEDLQTGPLKLEHLLAKGLTRKDLDLLKEGGLHTVECVAFAPLKALLAIKGISEQKAA 114
++ D L ++ L G+ DL LK GG++TV V + L IKG+SE K
Sbjct 7 EIDSDTAKNILSVDELQNYGINASDLQKLKSGGIYTVNTVLSTTRRHLCKIKGLSEVKVE 66
Query 115 KLKQASKELCSLGFCSAQEYLEAR 138
K+K+A+ ++ +GF A L+ R
Sbjct 67 KIKEAAGKIIQVGFIPATVQLDIR 90
> CE29064
Length=395
Score = 46.2 bits (108), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 22/62 (35%), Positives = 36/62 (58%), Gaps = 0/62 (0%)
Query 83 LKEGGLHTVECVAFAPLKALLAIKGISEQKAAKLKQASKELCSLGFCSAQEYLEARANLI 142
LKE G +T E +AF + L +KGIS+QKA K+ + + + +GF + E R+ L+
Sbjct 95 LKEAGYYTYESLAFTTRRELRNVKGISDQKAEKIMKEAMKFVQMGFTTGAEVHVKRSQLV 154
Query 143 KF 144
+
Sbjct 155 QI 156
> Hs5901996
Length=340
Score = 45.4 bits (106), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 25/72 (34%), Positives = 39/72 (54%), Gaps = 0/72 (0%)
Query 74 GLTRKDLDLLKEGGLHTVECVAFAPLKALLAIKGISEQKAAKLKQASKELCSLGFCSAQE 133
G+ D LK G+ T++ + +AL +KG+SE K K+K+A+ +L GF +A E
Sbjct 31 GINVADNKKLKSVGICTIKGIQMTTRRALCNVKGLSEAKVDKIKEAANKLIEPGFLTAFE 90
Query 134 YLEARANLIKFT 145
Y E R + T
Sbjct 91 YSEKRKMVFHIT 102
> Hs18254478
Length=217
Score = 30.0 bits (66), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 18/52 (34%), Positives = 26/52 (50%), Gaps = 0/52 (0%)
Query 53 LQDVGEDLQTGPLKLEHLLAKGLTRKDLDLLKEGGLHTVECVAFAPLKALLA 104
+QD+ DLQ G +K LL DLD LK+ + + VA+ A L+
Sbjct 57 VQDLSSDLQVGVIKPWLLLGSQDAAHDLDTLKKNKVTHILNVAYGVENAFLS 108
> Hs18547550
Length=138
Score = 29.6 bits (65), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 11/27 (40%), Positives = 18/27 (66%), Gaps = 0/27 (0%)
Query 78 KDLDLLKEGGLHTVECVAFAPLKALLA 104
D++L +E LHT+ C+A +P LL+
Sbjct 111 SDINLFREPFLHTLSCIAASPTSCLLS 137
> At4g02410
Length=674
Score = 29.6 bits (65), Expect = 1.9, Method: Composition-based stats.
Identities = 27/120 (22%), Positives = 48/120 (40%), Gaps = 1/120 (0%)
Query 8 SSLRRHRRGGGLPSLCFYLWPTAHGLDFRPMSSTVQETRQAPEQQLQDVGEDLQTGPLKL 67
+SL+R + +P L P + + + R+ ++ +D + L+
Sbjct 287 TSLQRFYKNR-MPLFSLLLIPVLFVVSLIFLVRFIVRRRRKFAEEFEDWETEFGKNRLRF 345
Query 68 EHLLAKGLTRKDLDLLKEGGLHTVECVAFAPLKALLAIKGISEQKAAKLKQASKELCSLG 127
+ L KD DLL GG V K +A+K +S + LK+ E+ S+G
Sbjct 346 KDLYYATKGFKDKDLLGSGGFGRVYRGVMPTTKKEIAVKRVSNESRQGLKEFVAEIVSIG 405
> Hs18547552
Length=138
Score = 29.6 bits (65), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 11/27 (40%), Positives = 18/27 (66%), Gaps = 0/27 (0%)
Query 78 KDLDLLKEGGLHTVECVAFAPLKALLA 104
D++L +E LHT+ C+A +P LL+
Sbjct 111 SDINLFREPFLHTLSCIAASPTSCLLS 137
> Hs17463795
Length=255
Score = 28.9 bits (63), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 33/109 (30%), Positives = 44/109 (40%), Gaps = 14/109 (12%)
Query 38 MSSTVQETRQAPEQQLQDVGEDLQTGP---------LKLEHLLAKGLTRKDLDLLKEGGL 88
M VQE P Q +D+ EDLQ P L+LEH + ++ G
Sbjct 12 MPLRVQEAAVEPMQVDKDLPEDLQNAPDINYLVDATLELEHSAKHAINMSSTSSSEDRGA 71
Query 89 HTVECVAFAPLKALLAIKGISEQKAAKLKQASK--ELCSLGFCSAQEYL 135
+ A A L L G++E A K KQA+K L S C + L
Sbjct 72 AS---QAQAVLTQLQCAGGLAELVARKYKQAAKCFLLASFDHCDVPQLL 117
> CE21056
Length=494
Score = 28.5 bits (62), Expect = 4.1, Method: Composition-based stats.
Identities = 14/37 (37%), Positives = 18/37 (48%), Gaps = 8/37 (21%)
Query 2 WLYLLQ-SSLRRHRRG-------GGLPSLCFYLWPTA 30
WL + Q + RH G G LP +C+YLW T
Sbjct 13 WLIVRQYQKVSRHPPGPISFPLIGNLPQICYYLWSTG 49
> At4g02420
Length=669
Score = 28.5 bits (62), Expect = 4.9, Method: Composition-based stats.
Identities = 19/86 (22%), Positives = 39/86 (45%), Gaps = 0/86 (0%)
Query 42 VQETRQAPEQQLQDVGEDLQTGPLKLEHLLAKGLTRKDLDLLKEGGLHTVECVAFAPLKA 101
+ + R+ ++++D + L+ + L KD ++L GG +V K
Sbjct 315 IMKRRRKFAEEVEDWETEFGKNRLRFKDLYYATKGFKDKNILGSGGFGSVYKGIMPKTKK 374
Query 102 LLAIKGISEQKAAKLKQASKELCSLG 127
+A+K +S + LK+ E+ S+G
Sbjct 375 EIAVKRVSNESRQGLKEFVAEIVSIG 400
> 7292573
Length=1280
Score = 28.1 bits (61), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 15/61 (24%), Positives = 32/61 (52%), Gaps = 1/61 (1%)
Query 2 WLYLLQSSLRRHRRGGGLPSLCFYLWPTAHGLDFRPMSSTVQETRQAPEQQLQDVGEDLQ 61
W+ + R+ R G +P +C +L+ T++ + ++ V E ++ P +Q+ + LQ
Sbjct 848 WVSRANAKQRKGRAGRVMPGVCIHLY-TSYRYQYHILAQPVPEIQRVPLEQIVLRIKTLQ 906
Query 62 T 62
T
Sbjct 907 T 907
Lambda K H
0.321 0.136 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1712413322
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40