bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0390_orf1
Length=163
Score E
Sequences producing significant alignments: (Bits) Value
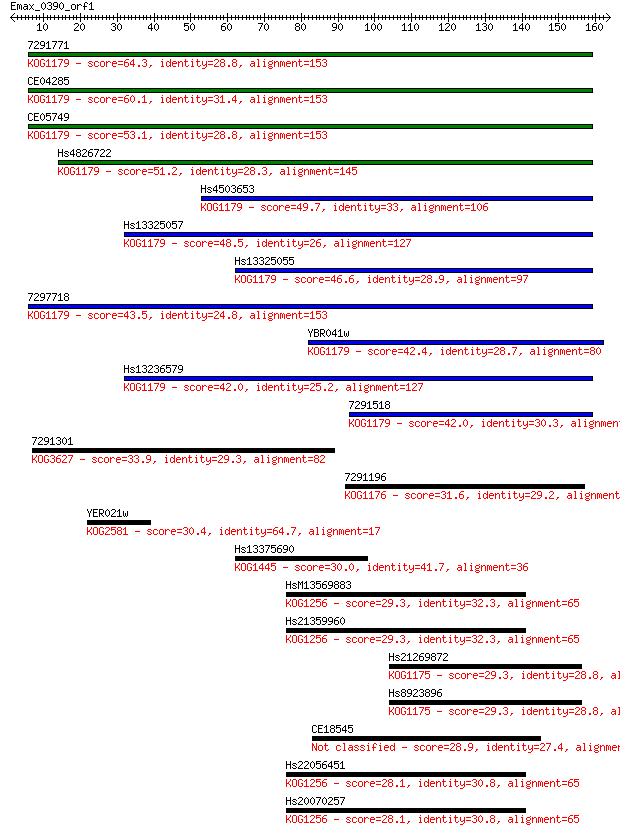
7291771 64.3 9e-11
CE04285 60.1 2e-09
CE05749 53.1 2e-07
Hs4826722 51.2 9e-07
Hs4503653 49.7 3e-06
Hs13325057 48.5 5e-06
Hs13325055 46.6 2e-05
7297718 43.5 2e-04
YBR041w 42.4 4e-04
Hs13236579 42.0 5e-04
7291518 42.0 5e-04
7291301 33.9 0.14
7291196 31.6 0.76
YER021w 30.4 1.5
Hs13375690 30.0 2.1
HsM13569883 29.3 3.2
Hs21359960 29.3 3.2
Hs21269872 29.3 3.2
Hs8923896 29.3 3.6
CE18545 28.9 4.7
Hs22056451 28.1 7.2
Hs20070257 28.1 7.6
> 7291771
Length=661
Score = 64.3 bits (155), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 44/156 (28%), Positives = 73/156 (46%), Gaps = 10/156 (6%)
Query 6 LFNAWNMPGACAFVPDFAWNSKELERIVEFNEETNEVYRDPTTKRGKEASRDEKTLVQRG 65
L N N GA FVP + + ++ ++ +E T E+ +D R + Q G
Sbjct 412 LINITNRVGAIGFVPVYGSSLYPVQ-VLRCDEYTGELLKDSK----GHCIRCQPG--QAG 464
Query 66 ELVSHIDPEHGLCLFTNY---GETEQFLYRDLLEAGDLWCRSGDIMERDAAGFLYLSKRI 122
LV +D + F Y G +EQ L R++ +GD++ SGD++ RD G+ Y R
Sbjct 465 LLVGKVDARRAVSAFHGYADKGASEQKLLRNVFTSGDVFFNSGDMVVRDILGYFYFKDRT 524
Query 123 GSSFFIDGEAAPVHELAKSLEHESGVREAHVEGLQV 158
G +F GE E+ + + G+ + V G+Q+
Sbjct 525 GDTFRWRGENVATQEVEAIITNCVGLEDCVVYGVQI 560
> CE04285
Length=655
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 48/157 (30%), Positives = 71/157 (45%), Gaps = 10/157 (6%)
Query 6 LFNAWNMPGACAFVPDFA-WNSKELERIVEFNEETNEVYRDPTTKRGKEASRDEKTLVQR 64
+ N N GAC F+P + S R+++ + T E+ RD K +
Sbjct 398 IVNVDNHVGACGFMPIYPHIGSLYPVRLIKVDRATGELERD------KNGLCVPCVPGET 451
Query 65 GELVSHIDPEHGLCLFTNY---GETEQFLYRDLLEAGDLWCRSGDIMERDAAGFLYLSKR 121
GE+V I + L F Y G+T + +YRD+ + GD SGDI+ D G+LY R
Sbjct 452 GEMVGVIKEKDILLKFEGYVSEGDTAKKIYRDVFKHGDKVFASGDILHWDDLGYLYFVDR 511
Query 122 IGSSFFIDGEAAPVHELAKSLEHESGVREAHVEGLQV 158
G +F GE E+ L+ V +A V G+ V
Sbjct 512 CGDTFRWKGENVSTTEVEGILQPVMDVEDATVYGVTV 548
> CE05749
Length=650
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 44/157 (28%), Positives = 70/157 (44%), Gaps = 10/157 (6%)
Query 6 LFNAWNMPGACAFVPDFAWNSK-ELERIVEFNEETNEVYRDPTTKRGKEASRDEKTLVQR 64
L N GAC F+P K R+++ ++ T E R T G + + +
Sbjct 394 LVNIDGHVGACGFLPISPLTKKMHPVRLIKVDDVTGEAIR---TSDGLCIACNPG---ES 447
Query 65 GELVSHIDPEHGLCLFTNY---GETEQFLYRDLLEAGDLWCRSGDIMERDAAGFLYLSKR 121
G +VS I + L F Y ET + + RD+ GD +GD++ D G++Y R
Sbjct 448 GAMVSTIRKNNPLLQFEGYLNKKETNKKIIRDVFAKGDSCFLTGDLLHWDRLGYVYFKDR 507
Query 122 IGSSFFIDGEAAPVHELAKSLEHESGVREAHVEGLQV 158
G +F GE E+ L +G+ +A V G++V
Sbjct 508 TGDTFRWKGENVSTTEVEAILHPITGLSDATVYGVEV 544
> Hs4826722
Length=641
Score = 51.2 bits (121), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 41/150 (27%), Positives = 64/150 (42%), Gaps = 14/150 (9%)
Query 14 GACAFVPDFAWNSKELE-----RIVEFNEETNEVYRDPTTKRGKEASRDEKTLVQRGELV 68
GAC F NS+ L R+V NE+T E+ R P + LV R +
Sbjct 400 GACGF------NSRILSFVYPIRLVRVNEDTMELIRGPDGVCIPCQPGEPGQLVGR---I 450
Query 69 SHIDPEHGLCLFTNYGETEQFLYRDLLEAGDLWCRSGDIMERDAAGFLYLSKRIGSSFFI 128
DP + N G + + +D+ + GD +GD++ D G+LY R G +F
Sbjct 451 IQKDPLRRFDGYLNQGANNKKIAKDVFKKGDQAYLTGDVLVMDELGYLYFRDRTGDTFRW 510
Query 129 DGEAAPVHELAKSLEHESGVREAHVEGLQV 158
GE E+ +L + + V G++V
Sbjct 511 KGENVSTTEVEGTLSRLLDMADVAVYGVEV 540
> Hs4503653
Length=620
Score = 49.7 bits (117), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 35/113 (30%), Positives = 54/113 (47%), Gaps = 7/113 (6%)
Query 53 EASRDEK---TLVQRGELVSHIDPEHGLCLFTNYG----ETEQFLYRDLLEAGDLWCRSG 105
E RDE V +GE+ + L F Y +TE+ RD+ + GDL+ SG
Sbjct 405 EPVRDENGYCVRVPKGEVGLLVCKITQLTPFNGYAGAKAQTEKKKLRDVFKKGDLYFNSG 464
Query 106 DIMERDAAGFLYLSKRIGSSFFIDGEAAPVHELAKSLEHESGVREAHVEGLQV 158
D++ D F+Y R+G +F GE E+A ++ V+E +V G+ V
Sbjct 465 DLLMVDHENFIYFHDRVGDTFRWKGENVATTEVADTVGLVDFVQEVNVYGVHV 517
> Hs13325057
Length=690
Score = 48.5 bits (114), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 33/128 (25%), Positives = 62/128 (48%), Gaps = 7/128 (5%)
Query 32 IVEFNEETNEVYRDPTTKRGKEASRDEKTLVQRGELVSHIDPEHGLCLFTNYGE-TEQFL 90
+V+F+ E E RD + L + G L++ + + + E +E+ L
Sbjct 466 LVQFDMEAAEPVRD------NQGFCIPVGLGEPGLLLTKVVSQQPFVGYRGPRELSERKL 519
Query 91 YRDLLEAGDLWCRSGDIMERDAAGFLYLSKRIGSSFFIDGEAAPVHELAKSLEHESGVRE 150
R++ ++GD++ +GD++ D GFLY R+G +F GE HE+ L +++
Sbjct 520 VRNVRQSGDVYYNTGDVLAMDREGFLYFRDRLGDTFRWKGENVSTHEVEGVLSQVDFLQQ 579
Query 151 AHVEGLQV 158
+V G+ V
Sbjct 580 VNVYGVCV 587
> Hs13325055
Length=619
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 28/101 (27%), Positives = 53/101 (52%), Gaps = 4/101 (3%)
Query 62 VQRGE---LVSHIDPEHGLCLFTN-YGETEQFLYRDLLEAGDLWCRSGDIMERDAAGFLY 117
V++GE L+S ++ ++ + Y T+ L D+ + GD++ +GD++ +D FLY
Sbjct 416 VKKGEPGLLISRVNAKNPFFGYAGPYKHTKDKLLCDVFKKGDVYLNTGDLIVQDQDNFLY 475
Query 118 LSKRIGSSFFIDGEAAPVHELAKSLEHESGVREAHVEGLQV 158
R G +F GE E+A + ++EA+V G+ +
Sbjct 476 FWDRTGDTFRWKGENVATTEVADVIGMLDFIQEANVYGVAI 516
> 7297718
Length=690
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 38/162 (23%), Positives = 65/162 (40%), Gaps = 22/162 (13%)
Query 6 LFNAWNMPGACAFVPDFAWNSKELERIVEFNEETNEVYRDPTTKRGKEASRDEKTLVQR- 64
+ N N GA FV RI+ + + DP T E RD L Q
Sbjct 441 IMNHDNTVGAIGFV----------SRILPKIYPISIIRADPDTG---EPIRDRNGLCQLC 487
Query 65 -----GELVSHI---DPEHGLCLFTNYGETEQFLYRDLLEAGDLWCRSGDIMERDAAGFL 116
G + I +P + + + + + +D+ + GD+ SGD++ D G+L
Sbjct 488 APNEPGVFIGKIVKGNPSREFLGYVDEKASAKKIVKDVFKHGDMAFISGDLLVADEKGYL 547
Query 117 YLSKRIGSSFFIDGEAAPVHELAKSLEHESGVREAHVEGLQV 158
Y R G +F GE E+ + + +G ++ V G+ +
Sbjct 548 YFKDRTGDTFRWKGENVSTSEVEAQVSNVAGYKDTVVYGVTI 589
> YBR041w
Length=623
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 23/80 (28%), Positives = 37/80 (46%), Gaps = 0/80 (0%)
Query 82 NYGETEQFLYRDLLEAGDLWCRSGDIMERDAAGFLYLSKRIGSSFFIDGEAAPVHELAKS 141
N ET+ + RD+ GD W R GD+++ D G Y R+G +F E E+
Sbjct 484 NAKETKSKVVRDVFRRGDAWYRCGDLLKADEYGLWYFLDRMGDTFRWKSENVSTTEVEDQ 543
Query 142 LEHESGVREAHVEGLQVFLP 161
L + + A V + + +P
Sbjct 544 LTASNKEQYAQVLVVGIKVP 563
> Hs13236579
Length=730
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 32/128 (25%), Positives = 57/128 (44%), Gaps = 7/128 (5%)
Query 32 IVEFNEETNEVYRDPTTKRGKEASRDEKTLVQRGELVSHIDPEHGLCLFTNYGETEQF-L 90
++ ++ T E RDP + + + G LV+ + + + E Q L
Sbjct 506 LIRYDVTTGEPIRDP------QGHCMATSPGEPGLLVAPVSQQSPFLGYAGGPELAQGKL 559
Query 91 YRDLLEAGDLWCRSGDIMERDAAGFLYLSKRIGSSFFIDGEAAPVHELAKSLEHESGVRE 150
+D+ GD++ +GD++ D GFL R G +F GE E+A+ E ++E
Sbjct 560 LKDVFRPGDVFFNTGDLLVCDDQGFLRFHDRTGDTFRWKGENVATTEVAEVFEALDFLQE 619
Query 151 AHVEGLQV 158
+V G+ V
Sbjct 620 VNVYGVTV 627
> 7291518
Length=664
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 20/66 (30%), Positives = 33/66 (50%), Gaps = 0/66 (0%)
Query 93 DLLEAGDLWCRSGDIMERDAAGFLYLSKRIGSSFFIDGEAAPVHELAKSLEHESGVREAH 152
D+ GD+ SGD++ D G+LY R G +F GE E+ L + +G ++
Sbjct 505 DVFSKGDMAFISGDLLVADERGYLYFKDRTGDTFRWKGENVSTSEVEAQLSNLAGYKDVI 564
Query 153 VEGLQV 158
V G+ +
Sbjct 565 VYGVSI 570
> 7291301
Length=709
Score = 33.9 bits (76), Expect = 0.14, Method: Composition-based stats.
Identities = 24/94 (25%), Positives = 40/94 (42%), Gaps = 23/94 (24%)
Query 7 FNAWNMPGACAFVPDFAWNSKELERIVEFNEETNEVYRDPTTKRGKEASRDEKTLVQRGE 66
F N+ C + D+ W+S L+ +V+FN TT G+ SR ++Q+
Sbjct 384 FTPNNIRSICLLMGDYTWSSNILKNLVKFN----------TTGWGRTESRINSPVLQQAS 433
Query 67 LVSH------------IDPEHGLCLFTNYGETEQ 88
L H +D H +C+ ++ G T Q
Sbjct 434 LTHHHLSYCAQVFGKQLDKSH-ICVASSTGSTCQ 466
> 7291196
Length=543
Score = 31.6 bits (70), Expect = 0.76, Method: Compositional matrix adjust.
Identities = 19/65 (29%), Positives = 31/65 (47%), Gaps = 0/65 (0%)
Query 92 RDLLEAGDLWCRSGDIMERDAAGFLYLSKRIGSSFFIDGEAAPVHELAKSLEHESGVREA 151
R L + +W R+GDI D+ G+LY+ R F + ++ + + GV EA
Sbjct 401 RRALSSDGMWFRTGDIGYLDSEGYLYIQTRDTDVFKFNNFQIYPEQIEEFILRLPGVSEA 460
Query 152 HVEGL 156
V G+
Sbjct 461 CVFGI 465
> YER021w
Length=523
Score = 30.4 bits (67), Expect = 1.5, Method: Composition-based stats.
Identities = 11/17 (64%), Positives = 15/17 (88%), Gaps = 0/17 (0%)
Query 22 FAWNSKELERIVEFNEE 38
F W+SKELE++VEFN +
Sbjct 145 FLWDSKELEQLVEFNRK 161
> Hs13375690
Length=925
Score = 30.0 bits (66), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 15/36 (41%), Positives = 19/36 (52%), Gaps = 0/36 (0%)
Query 62 VQRGELVSHIDPEHGLCLFTNYGETEQFLYRDLLEA 97
V L+ DP+ GL L T G+T FLY L E+
Sbjct 729 VAPSTLLPSYDPDTGLVLLTGKGDTRVFLYELLPES 764
> HsM13569883
Length=666
Score = 29.3 bits (64), Expect = 3.2, Method: Composition-based stats.
Identities = 21/69 (30%), Positives = 29/69 (42%), Gaps = 6/69 (8%)
Query 76 GLCLFTNYGETEQFLYRDLLEAGDLWCRSGDIMERDAAGFLYLSKRIGSSFFIDG----E 131
G +F Y E+E + + G W SGD+ + D GFLY++ I G
Sbjct 468 GRHIFMGYLESETETTEAIDDEG--WLHSGDLGQLDGLGFLYVTGHIKEILITAGGENVP 525
Query 132 AAPVHELAK 140
PV L K
Sbjct 526 PIPVETLVK 534
> Hs21359960
Length=666
Score = 29.3 bits (64), Expect = 3.2, Method: Composition-based stats.
Identities = 21/69 (30%), Positives = 29/69 (42%), Gaps = 6/69 (8%)
Query 76 GLCLFTNYGETEQFLYRDLLEAGDLWCRSGDIMERDAAGFLYLSKRIGSSFFIDG----E 131
G +F Y E+E + + G W SGD+ + D GFLY++ I G
Sbjct 468 GRHIFMGYLESETETTEAIDDEG--WLHSGDLGQLDGLGFLYVTGHIKEILITAGGENVP 525
Query 132 AAPVHELAK 140
PV L K
Sbjct 526 PIPVETLVK 534
> Hs21269872
Length=606
Score = 29.3 bits (64), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 15/52 (28%), Positives = 24/52 (46%), Gaps = 0/52 (0%)
Query 104 SGDIMERDAAGFLYLSKRIGSSFFIDGEAAPVHELAKSLEHESGVREAHVEG 155
+GD +RD G+ +++ RI + G E+ +L V EA V G
Sbjct 455 TGDGCQRDQDGYYWITGRIDDMLNVSGHLLSTAEVESALVEHEAVAEAAVVG 506
> Hs8923896
Length=701
Score = 29.3 bits (64), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 15/52 (28%), Positives = 24/52 (46%), Gaps = 0/52 (0%)
Query 104 SGDIMERDAAGFLYLSKRIGSSFFIDGEAAPVHELAKSLEHESGVREAHVEG 155
+GD +RD G+ +++ RI + G E+ +L V EA V G
Sbjct 550 TGDGCQRDQDGYYWITGRIDDMLNVSGHLLSTAEVESALVEHEAVAEAAVVG 601
> CE18545
Length=738
Score = 28.9 bits (63), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 17/62 (27%), Positives = 28/62 (45%), Gaps = 0/62 (0%)
Query 83 YGETEQFLYRDLLEAGDLWCRSGDIMERDAAGFLYLSKRIGSSFFIDGEAAPVHELAKSL 142
Y E E + LLE ++ CR + E+ A + Y+ + + S F D P + +
Sbjct 367 YLEMELHPHGPLLEPPEIRCRMIKMFEKHWAKYRYVFRDLHSYMFSDALGLPPKYIRQRF 426
Query 143 EH 144
EH
Sbjct 427 EH 428
> Hs22056451
Length=516
Score = 28.1 bits (61), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 20/69 (28%), Positives = 27/69 (39%), Gaps = 6/69 (8%)
Query 76 GLCLFTNYGETEQFLYRDLLEAGDLWCRSGDIMERDAAGFLYLSKRIGSSFFIDG----E 131
G +F Y E + E G W +GD DA GFLY++ R+ G
Sbjct 314 GRTIFMGYLNMEDKTCEAIDEEG--WLHTGDAGRLDADGFLYITGRLKELIITAGGENVP 371
Query 132 AAPVHELAK 140
P+ E K
Sbjct 372 PVPIEEAVK 380
> Hs20070257
Length=724
Score = 28.1 bits (61), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 20/69 (28%), Positives = 27/69 (39%), Gaps = 6/69 (8%)
Query 76 GLCLFTNYGETEQFLYRDLLEAGDLWCRSGDIMERDAAGFLYLSKRIGSSFFIDG----E 131
G +F Y E + E G W +GD DA GFLY++ R+ G
Sbjct 522 GRTIFMGYLNMEDKTCEAIDEEG--WLHTGDAGRLDADGFLYITGRLKELIITAGGENVP 579
Query 132 AAPVHELAK 140
P+ E K
Sbjct 580 PVPIEEAVK 588
Lambda K H
0.318 0.137 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2281134618
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40