bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
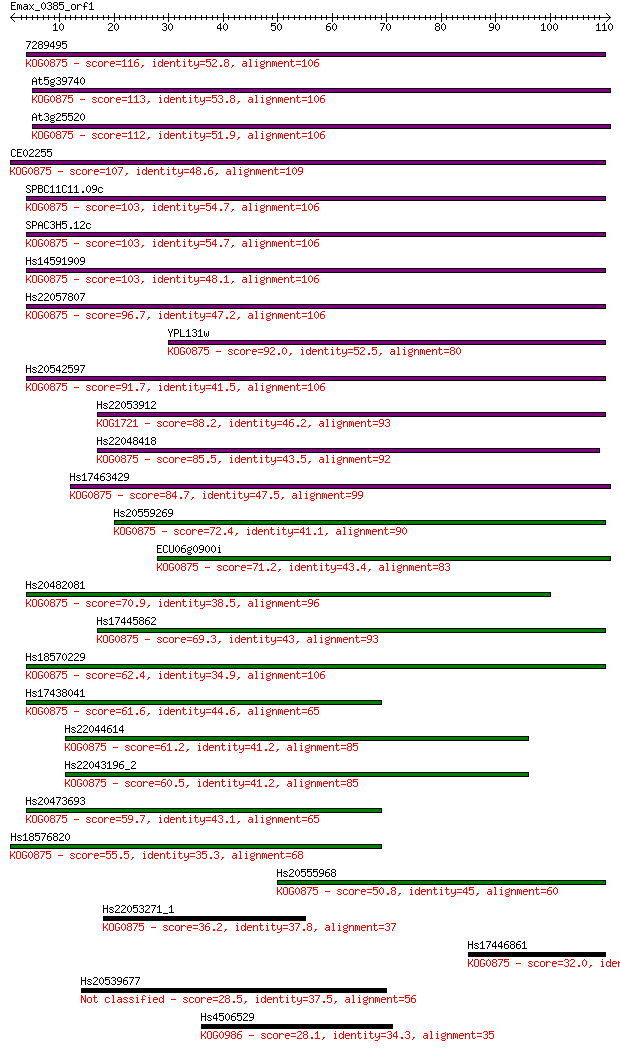
Query= Emax_0385_orf1
Length=110
Score E
Sequences producing significant alignments: (Bits) Value
7289495 116 9e-27
At5g39740 113 1e-25
At3g25520 112 2e-25
CE02255 107 8e-24
SPBC11C11.09c 103 6e-23
SPAC3H5.12c 103 6e-23
Hs14591909 103 7e-23
Hs22057807 96.7 1e-20
YPL131w 92.0 2e-19
Hs20542597 91.7 3e-19
Hs22053912 88.2 3e-18
Hs22048418 85.5 2e-17
Hs17463429 84.7 4e-17
Hs20559269 72.4 2e-13
ECU06g0900i 71.2 4e-13
Hs20482081 70.9 6e-13
Hs17445862 69.3 2e-12
Hs18570229 62.4 2e-10
Hs17438041 61.6 3e-10
Hs22044614 61.2 4e-10
Hs22043196_2 60.5 7e-10
Hs20473693 59.7 1e-09
Hs18576820 55.5 3e-08
Hs20555968 50.8 7e-07
Hs22053271_1 36.2 0.016
Hs17446861 32.0 0.27
Hs20539677 28.5 3.1
Hs4506529 28.1 4.5
> 7289495
Length=319
Score = 116 bits (291), Expect = 9e-27, Method: Compositional matrix adjust.
Identities = 56/106 (52%), Positives = 73/106 (68%), Gaps = 4/106 (3%)
Query 4 GLADEFVGVEKPNGEEFHIEETSEERRPFKCVLDVGIVSTTVGNRVFGAMKGACDGGLHI 63
GL + G + GEEF++E + F+C LDVG+ TT G RVFGAMKGA DGGL+I
Sbjct 134 GLDSLYAGCTEVTGEEFNVEPVDDGPGAFRCFLDVGLARTTTGARVFGAMKGAVDGGLNI 193
Query 64 PHSNKRFPGFTKGEDGAEDSYNPEVHRSRIYGLHVAEYMRTLKEED 109
PHS KRFPG++ S+N +VHR+ I+G HVA+YMR+L+EED
Sbjct 194 PHSVKRFPGYS----AETKSFNADVHRAHIFGQHVADYMRSLEEED 235
> At5g39740
Length=301
Score = 113 bits (282), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 57/106 (53%), Positives = 77/106 (72%), Gaps = 5/106 (4%)
Query 5 LADEFVGVEKPNGEEFHIEETSEERRPFKCVLDVGIVSTTVGNRVFGAMKGACDGGLHIP 64
+ DE+ G + GE+F +E T + RRPF+ +LDVG++ TT GNRVFGA+KGA DGGL IP
Sbjct 115 MDDEYEGNVEATGEDFSVEPT-DSRRPFRALLDVGLIRTTTGNRVFGALKGALDGGLDIP 173
Query 65 HSNKRFPGFTKGEDGAEDSYNPEVHRSRIYGLHVAEYMRTLKEEDP 110
HS+KRF GF K E+ D+ E+HR+ IYG HV+ YM+ L E++P
Sbjct 174 HSDKRFAGFHK-ENKQLDA---EIHRNYIYGGHVSNYMKLLGEDEP 215
> At3g25520
Length=190
Score = 112 bits (280), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 55/106 (51%), Positives = 74/106 (69%), Gaps = 5/106 (4%)
Query 5 LADEFVGVEKPNGEEFHIEETSEERRPFKCVLDVGIVSTTVGNRVFGAMKGACDGGLHIP 64
+ DE+ G + GE+F +E T + RRPF+ +LDVG++ TT GNRVFGA+KGA DGGL IP
Sbjct 4 MDDEYEGNVEATGEDFSVEPT-DSRRPFRALLDVGLIRTTTGNRVFGALKGALDGGLDIP 62
Query 65 HSNKRFPGFTKGEDGAEDSYNPEVHRSRIYGLHVAEYMRTLKEEDP 110
HS+KRF GF K + E+HR+ IYG HV+ YM+ L E++P
Sbjct 63 HSDKRFAGFHK----ENKQLDAEIHRNYIYGGHVSNYMKLLGEDEP 104
> CE02255
Length=293
Score = 107 bits (266), Expect = 8e-24, Method: Compositional matrix adjust.
Identities = 53/109 (48%), Positives = 69/109 (63%), Gaps = 5/109 (4%)
Query 1 KQKGLADEFVGVEKPNGEEFHIEETSEERRPFKCVLDVGIVSTTVGNRVFGAMKGACDGG 60
K GL + G E+ GE++++EE + R PFK VLD+G+ TT G+++F MKG DGG
Sbjct 111 KTIGLDSTYKGHEELTGEDYNVEEEGD-RAPFKAVLDIGLARTTTGSKIFAVMKGVADGG 169
Query 61 LHIPHSNKRFPGFTKGEDGAEDSYNPEVHRSRIYGLHVAEYMRTLKEED 109
+++PHS RF GF D YN E HR RI G HVA+YM LKEED
Sbjct 170 INVPHSESRFFGF----DQESKEYNAEAHRDRILGKHVADYMTYLKEED 214
> SPBC11C11.09c
Length=294
Score = 103 bits (258), Expect = 6e-23, Method: Compositional matrix adjust.
Identities = 58/107 (54%), Positives = 70/107 (65%), Gaps = 6/107 (5%)
Query 4 GLADEFVGVEKPNGEEFHIEETSEE-RRPFKCVLDVGIVSTTVGNRVFGAMKGACDGGLH 62
GLAD++ GV +P GE F + E E+ RPFK LDVG+ T+ G+RVFGAMKGA DGGL
Sbjct 114 GLADKYEGVTEPEGE-FELTEAIEDGPRPFKVFLDVGLKRTSTGSRVFGAMKGASDGGLF 172
Query 63 IPHSNKRFPGFTKGEDGAEDSYNPEVHRSRIYGLHVAEYMRTLKEED 109
IPHS RFPGF D + + E R IYG HVAEYM L ++D
Sbjct 173 IPHSPNRFPGF----DIETEELDDETLRKYIYGGHVAEYMEMLIDDD 215
> SPAC3H5.12c
Length=294
Score = 103 bits (258), Expect = 6e-23, Method: Compositional matrix adjust.
Identities = 58/107 (54%), Positives = 70/107 (65%), Gaps = 6/107 (5%)
Query 4 GLADEFVGVEKPNGEEFHIEETSEE-RRPFKCVLDVGIVSTTVGNRVFGAMKGACDGGLH 62
GLAD++ GV +P GE F + E E+ RPFK LDVG+ T+ G+RVFGAMKGA DGGL
Sbjct 114 GLADKYEGVTEPEGE-FELTEAIEDGPRPFKVFLDVGLKRTSTGSRVFGAMKGASDGGLF 172
Query 63 IPHSNKRFPGFTKGEDGAEDSYNPEVHRSRIYGLHVAEYMRTLKEED 109
IPHS RFPGF D + + E R IYG HVAEYM L ++D
Sbjct 173 IPHSPNRFPGF----DIETEELDDETLRKYIYGGHVAEYMEMLIDDD 215
> Hs14591909
Length=297
Score = 103 bits (257), Expect = 7e-23, Method: Compositional matrix adjust.
Identities = 51/106 (48%), Positives = 66/106 (62%), Gaps = 4/106 (3%)
Query 4 GLADEFVGVEKPNGEEFHIEETSEERRPFKCVLDVGIVSTTVGNRVFGAMKGACDGGLHI 63
G+ + G + G+E+++E + F C LD G+ TT GN+VFGA+KGA DGGL I
Sbjct 114 GMDKIYEGQVEVTGDEYNVESIDGQPGAFTCYLDAGLARTTTGNKVFGALKGAVDGGLSI 173
Query 64 PHSNKRFPGFTKGEDGAEDSYNPEVHRSRIYGLHVAEYMRTLKEED 109
PHS KRFPG+ D +N EVHR I G +VA+YMR L EED
Sbjct 174 PHSTKRFPGY----DSESKEFNAEVHRKHIMGQNVADYMRYLMEED 215
> Hs22057807
Length=247
Score = 96.7 bits (239), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 50/106 (47%), Positives = 66/106 (62%), Gaps = 4/106 (3%)
Query 4 GLADEFVGVEKPNGEEFHIEETSEERRPFKCVLDVGIVSTTVGNRVFGAMKGACDGGLHI 63
G+ + G + G+E+ +E + F C LD G+ TT GN+VFGA+KGA DGGL I
Sbjct 64 GMDKIYEGQVEVTGDEYIVESIDGQPSAFTCYLDAGLARTTTGNKVFGALKGAVDGGLSI 123
Query 64 PHSNKRFPGFTKGEDGAEDSYNPEVHRSRIYGLHVAEYMRTLKEED 109
PHS K+FPG+ G + E +N EVHR I G +VA+YM L EED
Sbjct 124 PHSTKQFPGY--GSESKE--FNAEVHRKHIMGQNVADYMCYLMEED 165
> YPL131w
Length=297
Score = 92.0 bits (227), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 42/80 (52%), Positives = 56/80 (70%), Gaps = 4/80 (5%)
Query 30 RPFKCVLDVGIVSTTVGNRVFGAMKGACDGGLHIPHSNKRFPGFTKGEDGAEDSYNPEVH 89
RPFK LD+G+ TT G RVFGA+KGA DGGL++PHS RFPG+ D + +PE+
Sbjct 140 RPFKVFLDIGLQRTTTGARVFGALKGASDGGLYVPHSENRFPGW----DFETEEIDPELL 195
Query 90 RSRIYGLHVAEYMRTLKEED 109
RS I+G HV++YM L ++D
Sbjct 196 RSYIFGGHVSQYMEELADDD 215
> Hs20542597
Length=336
Score = 91.7 bits (226), Expect = 3e-19, Method: Composition-based stats.
Identities = 44/106 (41%), Positives = 62/106 (58%), Gaps = 7/106 (6%)
Query 4 GLADEFVGVEKPNGEEFHIEETSEERRPFKCVLDVGIVSTTVGNRVFGAMKGACDGGLHI 63
G+ + + G + G+E+++E + F C LD G+ T GN+VFG +KG D L I
Sbjct 222 GMDNIYDGQMEVTGDEYNVENIDGQPGAFTCYLDAGLARTITGNKVFGTLKGTVDRSLSI 281
Query 64 PHSNKRFPGFTKGEDGAEDSYNPEVHRSRIYGLHVAEYMRTLKEED 109
PHS KRFPG+ + N EVH+ RI G +VA+YMR L E+D
Sbjct 282 PHSTKRFPGY-------DSESNAEVHQKRIMGQNVADYMRYLMEQD 320
> Hs22053912
Length=687
Score = 88.2 bits (217), Expect = 3e-18, Method: Composition-based stats.
Identities = 43/93 (46%), Positives = 59/93 (63%), Gaps = 4/93 (4%)
Query 17 GEEFHIEETSEERRPFKCVLDVGIVSTTVGNRVFGAMKGACDGGLHIPHSNKRFPGFTKG 76
G E+++E T + F C LDVG+ TT GN+VFG++K A DGGL IPHS K+FPG+
Sbjct 35 GNEYNVESTDGQPSAFTCYLDVGLARTTTGNKVFGSLKRAVDGGLSIPHSTKQFPGY--- 91
Query 77 EDGAEDSYNPEVHRSRIYGLHVAEYMRTLKEED 109
D +N VH+ I G +VA+YM L ++D
Sbjct 92 -DSESKDFNAAVHQKYIIGQNVADYMHYLMKDD 123
> Hs22048418
Length=200
Score = 85.5 bits (210), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 40/92 (43%), Positives = 55/92 (59%), Gaps = 4/92 (4%)
Query 17 GEEFHIEETSEERRPFKCVLDVGIVSTTVGNRVFGAMKGACDGGLHIPHSNKRFPGFTKG 76
G E+++E + F C LD G+ TT GN+VFGA+KGA +G L +PHS K+FPG+
Sbjct 33 GNEYNVESIDGQPGAFTCYLDTGLARTTTGNKVFGALKGAVNGSLSVPHSTKQFPGY--- 89
Query 77 EDGAEDSYNPEVHRSRIYGLHVAEYMRTLKEE 108
D +N EV R I +VAEY+ L E+
Sbjct 90 -DSESKEFNAEVQRKHIMDQNVAEYIHYLMED 120
> Hs17463429
Length=196
Score = 84.7 bits (208), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 47/101 (46%), Positives = 59/101 (58%), Gaps = 7/101 (6%)
Query 12 VEKPNGEEFHIEETSEERRPFKCVLDVGIVSTTVGNRVFGAMKGACDGGLHIPHSNKRFP 71
VEK G+E+++E + F LD + TT GN+VFGA+KG D GL IPHS KRFP
Sbjct 73 VEK-TGDEYNVESIDGQPGAFTYYLDASLARTTTGNQVFGALKGTVDEGLSIPHSTKRFP 131
Query 72 GFTKGEDGAEDSYNPEVHRSRIYGLHVAEYMRTLKE--EDP 110
G+ D +N EVH+ I G VA+YM L E EDP
Sbjct 132 GY----DSESKEFNAEVHQKYIMGQKVADYMHYLMEGDEDP 168
> Hs20559269
Length=127
Score = 72.4 bits (176), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 37/90 (41%), Positives = 51/90 (56%), Gaps = 15/90 (16%)
Query 20 FHIEETSEERRPFKCVLDVGIVSTTVGNRVFGAMKGACDGGLHIPHSNKRFPGFTKGEDG 79
+++E + F C LD G TT+GN+VFGA+KGA DGGL IPHS +
Sbjct 3 YNVESIDGQPGAFTCYLDAGFARTTIGNKVFGALKGAVDGGLSIPHSKE----------- 51
Query 80 AEDSYNPEVHRSRIYGLHVAEYMRTLKEED 109
+N EVH+ I G +VA++M L EE+
Sbjct 52 ----FNEEVHQKYIIGQNVADWMHHLMEEN 77
> ECU06g0900i
Length=283
Score = 71.2 bits (173), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 36/83 (43%), Positives = 56/83 (67%), Gaps = 5/83 (6%)
Query 28 ERRPFKCVLDVGIVSTTVGNRVFGAMKGACDGGLHIPHSNKRFPGFTKGEDGAEDSYNPE 87
E++ + LD+G+ +T G RVFGA+KGA D GL++PHS +F G DG+ D+ E
Sbjct 131 EKKAPRVFLDIGLARSTRGARVFGALKGASDAGLNVPHSAAKFYGHK--PDGSFDA--KE 186
Query 88 VHRSRIYGLHVAEYMRTLKEEDP 110
+H RI+G +++EYM+ L++ DP
Sbjct 187 LH-DRIFGYNISEYMKQLRDGDP 208
> Hs20482081
Length=195
Score = 70.9 bits (172), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 37/96 (38%), Positives = 52/96 (54%), Gaps = 4/96 (4%)
Query 4 GLADEFVGVEKPNGEEFHIEETSEERRPFKCVLDVGIVSTTVGNRVFGAMKGACDGGLHI 63
G+ + G + +E+++E + F C L G+ TT GN+VFG +K A GGL I
Sbjct 103 GMDKIYEGQVEATSDEYNVESIDGQPGAFTCYLGAGLARTTTGNKVFGTLKEAAVGGLSI 162
Query 64 PHSNKRFPGFTKGEDGAEDSYNPEVHRSRIYGLHVA 99
PHS K+FP + D +N EVHR I G +VA
Sbjct 163 PHSTKQFPDY----DSESKGFNAEVHRKDIMGQNVA 194
> Hs17445862
Length=146
Score = 69.3 bits (168), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 40/93 (43%), Positives = 50/93 (53%), Gaps = 9/93 (9%)
Query 17 GEEFHIEETSEERRPFKCVLDVGIVSTTVGNRVFGAMKGACDGGLHIPHSNKRFPGFTKG 76
G E+++E + F C L + TT GN+VFGA KGA DGGL PHS+KRF G
Sbjct 13 GNEYNVESIDGQPGAFTCYLVADLARTTTGNKVFGAPKGAVDGGLSNPHSSKRFLGL--- 69
Query 77 EDGAEDSYNPEVHRSRIYGLHVAEYMRTLKEED 109
S +VHR I G A+ +R L EED
Sbjct 70 ------SIPHKVHRKHIMGQKFADDLRYLIEED 96
> Hs18570229
Length=526
Score = 62.4 bits (150), Expect = 2e-10, Method: Composition-based stats.
Identities = 37/107 (34%), Positives = 54/107 (50%), Gaps = 7/107 (6%)
Query 4 GLADEFVGVEKPNGEEFHIEETSEERRPFKCVLDVGIVSTTVGNRVFGAMKGACDGGLHI 63
G+ + G + G+E+++E T + C LD G+ TT GN+VFG +KGA DGGL +
Sbjct 87 GMDKIYEGQVEVTGDEYNVESTDGQPGTLTCYLDAGLARTTTGNKVFGTLKGAVDGGLSV 146
Query 64 PH-SNKRFPGFTKGEDGAEDSYNPEVHRSRIYGLHVAEYMRTLKEED 109
PH N K + + Y R+R+ +YM L EED
Sbjct 147 PHIPNDSLVMILKARNLMQ-KYTGSTSRARML-----QYMCYLMEED 187
> Hs17438041
Length=209
Score = 61.6 bits (148), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 29/65 (44%), Positives = 39/65 (60%), Gaps = 0/65 (0%)
Query 4 GLADEFVGVEKPNGEEFHIEETSEERRPFKCVLDVGIVSTTVGNRVFGAMKGACDGGLHI 63
G+ + G + G+E+++E F C LD G+ TT GN+VFG +KGA DGGL I
Sbjct 145 GMDKIYEGQVEVTGDEYNVESIDGPPGAFTCYLDAGLARTTTGNKVFGTLKGAVDGGLSI 204
Query 64 PHSNK 68
PHS K
Sbjct 205 PHSTK 209
> Hs22044614
Length=455
Score = 61.2 bits (147), Expect = 4e-10, Method: Composition-based stats.
Identities = 35/90 (38%), Positives = 52/90 (57%), Gaps = 6/90 (6%)
Query 11 GVEKPNGEEFHIEETSEERRPFKCVLDVGIVSTTVGNRVFGAMKGACDGGLHIPHSNKRF 70
G E+ G+E+ +E + F LD G+ TT ++VFGAM+ A DGGL IP S K+F
Sbjct 315 GQEEVTGDEYDVESIDGQPDAFTYYLDAGLARTTTDSKVFGAME-AVDGGLSIPQSTKQF 373
Query 71 PGF--TKGEDGAEDSY---NPEVHRSRIYG 95
PG+ E A+++ + E+H SR+ G
Sbjct 374 PGYDSESKEFNAQETVLTIHKELHNSRLGG 403
> Hs22043196_2
Length=415
Score = 60.5 bits (145), Expect = 7e-10, Method: Composition-based stats.
Identities = 35/90 (38%), Positives = 52/90 (57%), Gaps = 6/90 (6%)
Query 11 GVEKPNGEEFHIEETSEERRPFKCVLDVGIVSTTVGNRVFGAMKGACDGGLHIPHSNKRF 70
G E+ G+E+ +E + F LD G+ TT ++VFGAM+ A DGGL IP S K+F
Sbjct 275 GQEEVTGDEYDVESIDGQPDAFTYYLDAGLARTTTDSKVFGAME-AVDGGLSIPQSTKQF 333
Query 71 PGF--TKGEDGAEDSY---NPEVHRSRIYG 95
PG+ E A+++ + E+H SR+ G
Sbjct 334 PGYDSESKEFNAQETVLTIHKELHNSRLGG 363
> Hs20473693
Length=451
Score = 59.7 bits (143), Expect = 1e-09, Method: Composition-based stats.
Identities = 28/65 (43%), Positives = 38/65 (58%), Gaps = 0/65 (0%)
Query 4 GLADEFVGVEKPNGEEFHIEETSEERRPFKCVLDVGIVSTTVGNRVFGAMKGACDGGLHI 63
G+ + G K G+E+ +E + F C LD G+ TT GN+VFG +KGA DGGL I
Sbjct 387 GMDKIYEGQVKVTGDEYDVESIDGQPGAFTCYLDAGLARTTAGNQVFGTLKGAVDGGLSI 446
Query 64 PHSNK 68
P + K
Sbjct 447 PRNTK 451
> Hs18576820
Length=133
Score = 55.5 bits (132), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 24/68 (35%), Positives = 39/68 (57%), Gaps = 0/68 (0%)
Query 1 KQKGLADEFVGVEKPNGEEFHIEETSEERRPFKCVLDVGIVSTTVGNRVFGAMKGACDGG 60
+ G+ + G + G+E+++E T + F C LD G+ TT GN+V G ++G+ DGG
Sbjct 66 NRSGMGKIYEGQVEVTGDEYNVESTYGQPGAFTCYLDAGLAKTTSGNKVLGGLEGSTDGG 125
Query 61 LHIPHSNK 68
L PH +
Sbjct 126 LSTPHGTQ 133
> Hs20555968
Length=91
Score = 50.8 bits (120), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 27/60 (45%), Positives = 34/60 (56%), Gaps = 4/60 (6%)
Query 50 FGAMKGACDGGLHIPHSNKRFPGFTKGEDGAEDSYNPEVHRSRIYGLHVAEYMRTLKEED 109
+G KG DGGL IPH+ KRFPG+ D +N EVH+ I + A YM L E+D
Sbjct 27 YGVWKGVVDGGLLIPHNIKRFPGY----DSESKKFNAEVHQKPIMDPNAAGYMCYLIEKD 82
> Hs22053271_1
Length=329
Score = 36.2 bits (82), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 14/37 (37%), Positives = 23/37 (62%), Gaps = 0/37 (0%)
Query 18 EEFHIEETSEERRPFKCVLDVGIVSTTVGNRVFGAMK 54
+E+++ T + F C LD G+ TT N+VFGA++
Sbjct 200 DEYNVGSTDGQPGAFTCCLDAGLARTTTDNKVFGALR 236
> Hs17446861
Length=230
Score = 32.0 bits (71), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 15/25 (60%), Positives = 17/25 (68%), Gaps = 0/25 (0%)
Query 85 NPEVHRSRIYGLHVAEYMRTLKEED 109
PEVHR I G +VA+YM L EED
Sbjct 124 QPEVHRKHIMGHNVADYMCYLMEED 148
> Hs20539677
Length=260
Score = 28.5 bits (62), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 21/56 (37%), Positives = 28/56 (50%), Gaps = 3/56 (5%)
Query 14 KPNGEEFHIEETSEERRPFKCVLDVGIVSTTVGNRVFGAMKGACDGGLHIPHSNKR 69
K G++ + ET+ CV DV IV+T G+ F A+ D GL I HS R
Sbjct 168 KKQGQKAQVTETTVISHDILCVADVQIVTTYGGH--FCALFSVLDRGL-IWHSPLR 220
> Hs4506529
Length=563
Score = 28.1 bits (61), Expect = 4.5, Method: Composition-based stats.
Identities = 12/35 (34%), Positives = 19/35 (54%), Gaps = 0/35 (0%)
Query 36 LDVGIVSTTVGNRVFGAMKGACDGGLHIPHSNKRF 70
+D G + T V N F A +G+ DG P +K++
Sbjct 1 MDFGSLETVVANSAFIAARGSFDGSSSQPSRDKKY 35
Lambda K H
0.315 0.138 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1195973986
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40