bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0369_orf1
Length=151
Score E
Sequences producing significant alignments: (Bits) Value
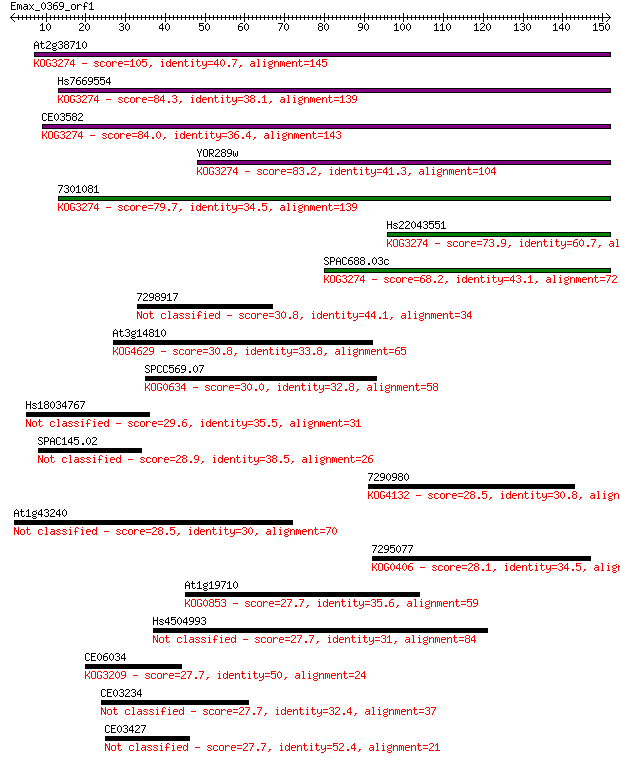
At2g38710 105 4e-23
Hs7669554 84.3 7e-17
CE03582 84.0 1e-16
YOR289w 83.2 2e-16
7301081 79.7 2e-15
Hs22043551 73.9 9e-14
SPAC688.03c 68.2 6e-12
7298917 30.8 0.99
At3g14810 30.8 1.0
SPCC569.07 30.0 1.8
Hs18034767 29.6 2.5
SPAC145.02 28.9 4.0
7290980 28.5 4.9
At1g43240 28.5 5.1
7295077 28.1 6.1
At1g19710 27.7 8.0
Hs4504993 27.7 8.6
CE06034 27.7 8.7
CE03234 27.7 9.1
CE03427 27.7 9.5
> At2g38710
Length=214
Score = 105 bits (262), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 59/146 (40%), Positives = 78/146 (53%), Gaps = 25/146 (17%)
Query 7 IEADESMCAWMFDTLIAHFDESNSSSPPPPPSIVSLHRLGVRTPAFVTWMKRRNSSSSSS 66
+ A+ M + FDTL++H+ N+ PPP + H P FVTW K N
Sbjct 2 VSANREMAVYCFDTLVSHY---NNEETPPPAFEEANH------PLFVTWKKIVNGGEPR- 51
Query 67 SSSSKEGFREDNVDLRGCIGSLKPIPIMK-LKDYALISALQDSRFSPITLSEIPDLKCHV 125
LRGCIG+L+ ++ KDYAL SAL+D RF PI E+P L+C V
Sbjct 52 --------------LRGCIGTLEARRLISGFKDYALTSALRDRRFPPIQAKELPSLQCTV 97
Query 126 SLLHSFERAKDVYDWEIGVHGIIIRF 151
S+L +E A+D DWE+G HGIII F
Sbjct 98 SVLTDYEDAEDYLDWEVGKHGIIIEF 123
> Hs7669554
Length=333
Score = 84.3 bits (207), Expect = 7e-17, Method: Compositional matrix adjust.
Identities = 53/140 (37%), Positives = 66/140 (47%), Gaps = 27/140 (19%)
Query 13 MCAWMFDTLIAHFDESNSSSPPPPPSIVSLHRLGVRTPAFVTWMKRRNSSSSSSSSSSKE 72
MC + FD L H P P + P FVTW R+
Sbjct 130 MCCFCFDVLYCHL---YGYQQPRTPRFTN-----EPYPLFVTWKIGRDKR---------- 171
Query 73 GFREDNVDLRGCIGSLKPIPIMK-LKDYALISALQDSRFSPITLSEIPDLKCHVSLLHSF 131
LRGCIG+ + + L++Y L SAL+DSRF P+T E+P L C VSLL +F
Sbjct 172 --------LRGCIGTFSAMNLHSGLREYTLTSALKDSRFPPMTRDELPRLFCSVSLLTNF 223
Query 132 ERAKDVYDWEIGVHGIIIRF 151
E D DWE+GVHGI I F
Sbjct 224 EDVCDYLDWEVGVHGIRIEF 243
> CE03582
Length=200
Score = 84.0 bits (206), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 52/144 (36%), Positives = 72/144 (50%), Gaps = 26/144 (18%)
Query 9 ADESMCAWMFDTLIAHFDESNSSSPPPPPSIVSLHRLGVRTPAFVTWMKRRNSSSSSSSS 68
A+ M + FD + A + PP P I + V+ P FVTW
Sbjct 4 ANIQMAVYCFDVINAQLNRE--KEPPVPKEIPN-----VKLPLFVTW------------- 43
Query 69 SSKEGFREDNVDLRGCIGSLKPIPIMK-LKDYALISALQDSRFSPITLSEIPDLKCHVSL 127
K+G + D LRGCIG+ + + + L +YA SA DSRF PI+ E+P L+C VSL
Sbjct 44 --KKGHQHD---LRGCIGTFSDLRLGEGLNEYAKTSAFHDSRFKPISREEVPSLQCGVSL 98
Query 128 LHSFERAKDVYDWEIGVHGIIIRF 151
L +FE + DW IG HG+ + F
Sbjct 99 LINFEPIHNFRDWTIGRHGVRMNF 122
> YOR289w
Length=251
Score = 83.2 bits (204), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 43/113 (38%), Positives = 66/113 (58%), Gaps = 16/113 (14%)
Query 48 RTPAFVTWMKRRNSSSSSSSSSSKEGFREDNVDLRGCIGSLKPIPIMK-LKDYALISALQ 106
+T F+TW K+ N + ++ E+N LRGCIG+ +PI ++ Y+LI+AL+
Sbjct 66 KTSLFITWKKKSNKHHTIDTN-------EENYILRGCIGTFAKMPIAHGIEKYSLIAALE 118
Query 107 DSRFSPITLSEIPDLKCHVSLLHSFER--------AKDVYDWEIGVHGIIIRF 151
D RFSPI E+ DLKC ++L +F+ D++DWE+G HGI + F
Sbjct 119 DRRFSPIQKRELVDLKCSCNILGNFKTIFRGGGNPNGDIFDWELGKHGIELYF 171
> 7301081
Length=243
Score = 79.7 bits (195), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 48/140 (34%), Positives = 68/140 (48%), Gaps = 27/140 (19%)
Query 13 MCAWMFDTLIAHFDESNSSSPPPPPSIVSLHRLGVRTPAFVTWMKRRNSSSSSSSSSSKE 72
MC + F+ L + + S P + P FVTW R+
Sbjct 49 MCLFCFEVLDCELNNVDGPSVPVFSNDA--------YPLFVTWKIGRDKR---------- 90
Query 73 GFREDNVDLRGCIGSLKPIPIMK-LKDYALISALQDSRFSPITLSEIPDLKCHVSLLHSF 131
LRGCIG+ + + L++YAL SA +DSRF+PI+ E+P L VS+L +F
Sbjct 91 --------LRGCIGTFSAMELHHGLREYALTSAFKDSRFAPISRDELPRLTVSVSILQNF 142
Query 132 ERAKDVYDWEIGVHGIIIRF 151
E A+ DW++GVHGI I F
Sbjct 143 EEAQGHLDWQLGVHGIRIEF 162
> Hs22043551
Length=151
Score = 73.9 bits (180), Expect = 9e-14, Method: Compositional matrix adjust.
Identities = 34/56 (60%), Positives = 41/56 (73%), Gaps = 0/56 (0%)
Query 96 LKDYALISALQDSRFSPITLSEIPDLKCHVSLLHSFERAKDVYDWEIGVHGIIIRF 151
L++Y L SAL+DSRF P+T E+P L C VSLL +FE A D DWE+GVHGI I F
Sbjct 7 LREYTLTSALKDSRFPPLTREELPKLFCSVSLLTNFEDASDYLDWEVGVHGIRIEF 62
> SPAC688.03c
Length=204
Score = 68.2 bits (165), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 31/73 (42%), Positives = 46/73 (63%), Gaps = 1/73 (1%)
Query 80 DLRGCIGSLKPIPIM-KLKDYALISALQDSRFSPITLSEIPDLKCHVSLLHSFERAKDVY 138
LRGCIG+ + P++ L ++ +A D RF PI+L E+ L+C + LL FE D
Sbjct 62 QLRGCIGTFRARPLVTNLTYFSKQAAFCDERFRPISLGELALLECQIDLLVDFEPIDDPL 121
Query 139 DWEIGVHGIIIRF 151
DWE+G+HG+ I+F
Sbjct 122 DWEVGIHGVSIKF 134
> 7298917
Length=2237
Score = 30.8 bits (68), Expect = 0.99, Method: Composition-based stats.
Identities = 15/35 (42%), Positives = 24/35 (68%), Gaps = 1/35 (2%)
Query 33 PPPPPSIVSLHRLGVR-TPAFVTWMKRRNSSSSSS 66
PPPP + SL+ G + TPA + + +RR+S S++S
Sbjct 1160 PPPPSHMASLYPAGQQTTPADLGYQRRRSSVSATS 1194
> At3g14810
Length=853
Score = 30.8 bits (68), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 22/75 (29%), Positives = 35/75 (46%), Gaps = 11/75 (14%)
Query 27 ESNSSSPPPPPSIVSLHRLG----------VRTPAFVTWMKRRNSSSSSSSSSSKEGFRE 76
E S++PP P + + R G V+T ++RR + +S SS +EG
Sbjct 115 ELQSNTPPRPATASNTPRRGLTTISESSSPVKTKVKADAVRRRQNRTSLGGSSDEEGRNR 174
Query 77 DNVDLRGCIGSLKPI 91
D ++ C GS KP+
Sbjct 175 DEAEVLKC-GSKKPM 188
> SPCC569.07
Length=470
Score = 30.0 bits (66), Expect = 1.8, Method: Composition-based stats.
Identities = 19/60 (31%), Positives = 28/60 (46%), Gaps = 3/60 (5%)
Query 35 PPPSIVSLHRLGVRTPAFVTWMKRRNSSSSSSSSSSKEGFREDNVDLRGCI--GSLKPIP 92
P PS +H+L V P +W K N ++ S S E ++DL G + G + IP
Sbjct 46 PNPSKFPIHKLSVSFPEVNSWEKDTNKDATVSYELSNNA-NEGSLDLLGALQYGQCQGIP 104
> Hs18034767
Length=704
Score = 29.6 bits (65), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 11/31 (35%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 5 GTIEADESMCAWMFDTLIAHFDESNSSSPPP 35
G+ EA+ C + D +++HF+ SSP P
Sbjct 548 GSFEANPPFCEELMDAMVSHFERLLESSPEP 578
> SPAC145.02
Length=270
Score = 28.9 bits (63), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 10/26 (38%), Positives = 17/26 (65%), Gaps = 0/26 (0%)
Query 8 EADESMCAWMFDTLIAHFDESNSSSP 33
+ D S+C W+ DT + F +SN++ P
Sbjct 80 KIDPSICKWILDTYLPLFKKSNNAYP 105
> 7290980
Length=277
Score = 28.5 bits (62), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 16/52 (30%), Positives = 27/52 (51%), Gaps = 1/52 (1%)
Query 91 IPIMKLKDYALISALQDSRFSPITLSEIPDLKCHVSLLHSFERAKDVYDWEI 142
+P L L+S L ++ FS + E+ + +CH L + ERA ++Y I
Sbjct 139 LPCGNLATDTLLSKLAENGFS-VDACEVYETRCHPELGANVERALEIYGESI 189
> At1g43240
Length=698
Score = 28.5 bits (62), Expect = 5.1, Method: Composition-based stats.
Identities = 21/72 (29%), Positives = 27/72 (37%), Gaps = 11/72 (15%)
Query 2 KHDGTIEADESMCAWMFDTLIAHFDESNSSSPPPPPSIVSL--HRLGVRTPAFVTWMKRR 59
KH GT+ S I H D PPP SI+S+ R+G+ W +R
Sbjct 322 KHTGTLRLIAS---------ILHDDYPGEFQTPPPKSIMSIVRGRMGLHCSYATAWRGKR 372
Query 60 NSSSSSSSSSSK 71
S S K
Sbjct 373 QHVSDVRGSPEK 384
> 7295077
Length=467
Score = 28.1 bits (61), Expect = 6.1, Method: Composition-based stats.
Identities = 19/59 (32%), Positives = 30/59 (50%), Gaps = 9/59 (15%)
Query 92 PIMKLKDYALISALQDSRFSPITLSEIPDLKCHVSL----LHSFERAKDVYDWEIGVHG 146
P+ K D LI RF+P+ + P L C+ + + +FE A DV++ E+G G
Sbjct 325 PLQKALDKILIE-----RFAPVVSAIYPVLTCNPNAPKDAIPNFENALDVFEVELGKRG 378
> At1g19710
Length=479
Score = 27.7 bits (60), Expect = 8.0, Method: Composition-based stats.
Identities = 21/61 (34%), Positives = 29/61 (47%), Gaps = 4/61 (6%)
Query 45 LGVRTPAFVTWMKRRNSSSSSSSSSSKEGFREDNVDLR--GCIGSLKPIPIMKLKDYALI 102
L V T + +S S S S+E +EDN D++ GSL P+ MK K L+
Sbjct 25 LSVSTVGMILVRSTFDSCSVSGKRCSRE--KEDNSDIKIQSVSGSLNPLEFMKSKLVLLV 82
Query 103 S 103
S
Sbjct 83 S 83
> Hs4504993
Length=1097
Score = 27.7 bits (60), Expect = 8.6, Method: Compositional matrix adjust.
Identities = 26/91 (28%), Positives = 39/91 (42%), Gaps = 9/91 (9%)
Query 37 PSIVSLHRLGVRTPAFVTWMKRRNS----SSSSSSSSSKEGFREDNVDLRGCIGSLKPIP 92
P + R+ T F W K N ++ +S S + +RE + D + I KP+P
Sbjct 501 PYTLYTFRIRCSTETFWKWSKWSNKKQHLTTEASPSKGPDTWREWSSDGKNLIIYWKPLP 560
Query 93 IMKLKDYAL---ISALQDSRFSPITLSEIPD 120
I + L +S D +LSEIPD
Sbjct 561 INEANGKILSYNVSCSSDEETQ--SLSEIPD 589
> CE06034
Length=1012
Score = 27.7 bits (60), Expect = 8.7, Method: Compositional matrix adjust.
Identities = 12/24 (50%), Positives = 15/24 (62%), Gaps = 1/24 (4%)
Query 20 TLIAHFDESNSSSPPPPPSIVSLH 43
T+ H+ ESN PPPPPS+ H
Sbjct 881 TMNGHY-ESNYGLPPPPPSVYEKH 903
> CE03234
Length=914
Score = 27.7 bits (60), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 12/37 (32%), Positives = 19/37 (51%), Gaps = 0/37 (0%)
Query 24 HFDESNSSSPPPPPSIVSLHRLGVRTPAFVTWMKRRN 60
H + SPP P S+ R+G+RT ++ +RN
Sbjct 449 HLSDVEVHSPPKTPQTPSILRVGIRTSSYPMTETKRN 485
> CE03427
Length=226
Score = 27.7 bits (60), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 11/21 (52%), Positives = 15/21 (71%), Gaps = 0/21 (0%)
Query 25 FDESNSSSPPPPPSIVSLHRL 45
FDE+N+ SP PPP+ + RL
Sbjct 39 FDEANTPSPAPPPTHIVRERL 59
Lambda K H
0.319 0.133 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1888713112
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40