bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0334_orf1
Length=148
Score E
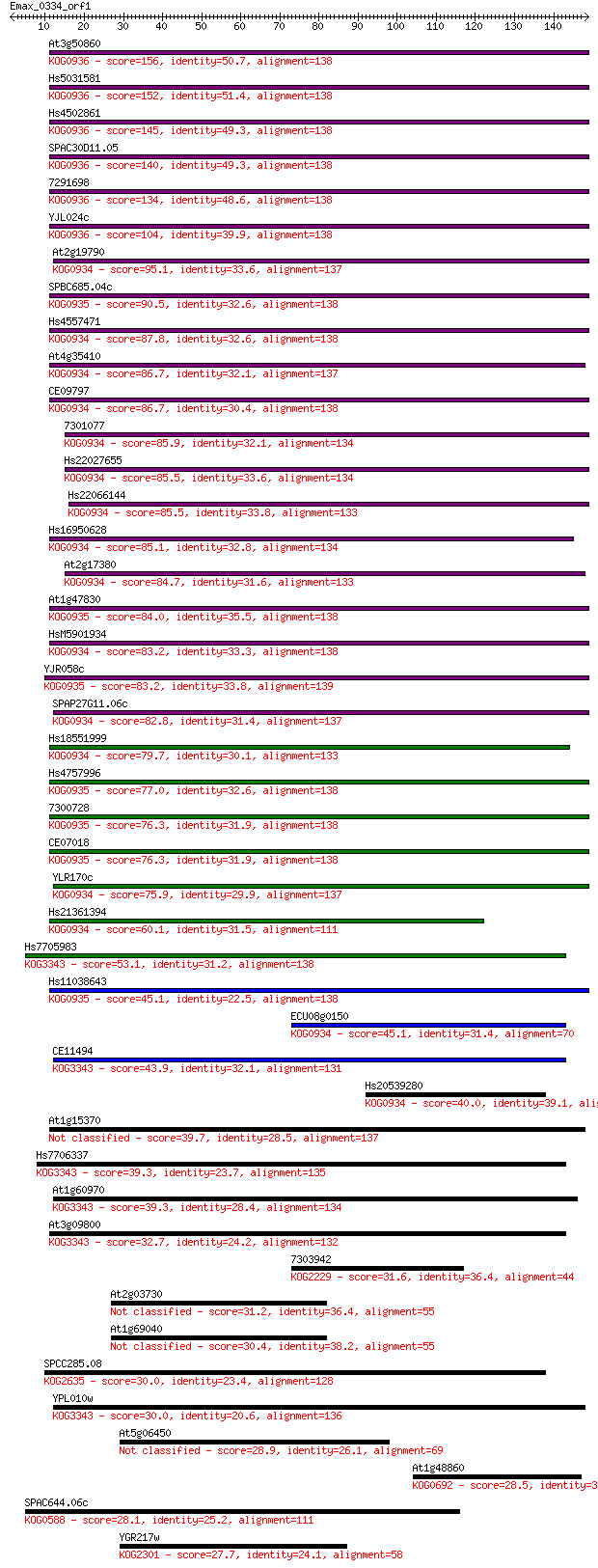
Sequences producing significant alignments: (Bits) Value
At3g50860 156 1e-38
Hs5031581 152 2e-37
Hs4502861 145 2e-35
SPAC30D11.05 140 1e-33
7291698 134 6e-32
YJL024c 104 7e-23
At2g19790 95.1 5e-20
SPBC685.04c 90.5 1e-18
Hs4557471 87.8 7e-18
At4g35410 86.7 1e-17
CE09797 86.7 2e-17
7301077 85.9 3e-17
Hs22027655 85.5 3e-17
Hs22066144 85.5 3e-17
Hs16950628 85.1 4e-17
At2g17380 84.7 6e-17
At1g47830 84.0 1e-16
HsM5901934 83.2 2e-16
YJR058c 83.2 2e-16
SPAP27G11.06c 82.8 2e-16
Hs18551999 79.7 2e-15
Hs4757996 77.0 1e-14
7300728 76.3 2e-14
CE07018 76.3 2e-14
YLR170c 75.9 3e-14
Hs21361394 60.1 1e-09
Hs7705983 53.1 2e-07
Hs11038643 45.1 5e-05
ECU08g0150 45.1 5e-05
CE11494 43.9 1e-04
Hs20539280 40.0 0.002
At1g15370 39.7 0.002
Hs7706337 39.3 0.002
At1g60970 39.3 0.003
At3g09800 32.7 0.24
7303942 31.6 0.60
At2g03730 31.2 0.69
At1g69040 30.4 1.3
SPCC285.08 30.0 1.6
YPL010w 30.0 1.6
At5g06450 28.9 3.9
At1g48860 28.5 4.5
SPAC644.06c 28.1 6.1
YGR217w 27.7 7.9
> At3g50860
Length=145
Score = 156 bits (395), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 70/138 (50%), Positives = 102/138 (73%), Gaps = 1/138 (0%)
Query 11 MIKSVVVVNTQGKPRIVRFYDGTPQEKHQHVMKSLFAAVSRRPSEGCCCFTENTDLFGPN 70
MIK+V+++NTQGKPR+ +FYD P EK Q +++ +F+ + RP E F E LFGP+
Sbjct 1 MIKAVMMMNTQGKPRLAKFYDYLPVEKQQELIRGVFSVLCSRP-ENVSNFLEIESLFGPD 59
Query 71 HRIIYRHFATLFFIFVTDNLESELGLLDLIQVVVQVLDACFENVCELDLVFHYDKINYIL 130
R++Y+H+ATL+F+ V D E+EL +LDLIQV+V+ LD CF NVCELD+VF+Y K++ +L
Sbjct 60 SRLVYKHYATLYFVLVFDGSENELAMLDLIQVLVETLDKCFSNVCELDIVFNYSKMHAVL 119
Query 131 DEIIVGGLVMETGVDNIL 148
DEI+ GG V+ET ++
Sbjct 120 DEIVFGGQVLETSSAEVM 137
> Hs5031581
Length=193
Score = 152 bits (385), Expect = 2e-37, Method: Compositional matrix adjust.
Identities = 71/139 (51%), Positives = 103/139 (74%), Gaps = 2/139 (1%)
Query 11 MIKSVVVVNTQGKPRIVRFYDGTPQEKHQHVMKSLFAAVSRRPSEGCCCFTENTDLFG-P 69
MI++++V N GKPR+VRFY P+E Q +++ F V +R + C F E L G
Sbjct 1 MIQAILVFNNHGKPRLVRFYQRFPEEIQQQIVRETFHLVLKR-DDNICNFLEGGSLIGGS 59
Query 70 NHRIIYRHFATLFFIFVTDNLESELGLLDLIQVVVQVLDACFENVCELDLVFHYDKINYI 129
++++IYRH+ATL+F+F D+ ESELG+LDLIQV V+ LD CFENVCELDL+FH DK++YI
Sbjct 60 DYKLIYRHYATLYFVFCVDSSESELGILDLIQVFVETLDKCFENVCELDLIFHMDKVHYI 119
Query 130 LDEIIVGGLVMETGVDNIL 148
L E+++GG+V+ET ++ I+
Sbjct 120 LQEVVMGGMVLETNMNEIV 138
> Hs4502861
Length=193
Score = 145 bits (367), Expect = 2e-35, Method: Compositional matrix adjust.
Identities = 68/139 (48%), Positives = 101/139 (72%), Gaps = 2/139 (1%)
Query 11 MIKSVVVVNTQGKPRIVRFYDGTPQEKHQHVMKSLFAAVSRRPSEGCCCFTENTDLFG-P 69
MIK++++ N GKPR+ +FY ++ Q +++ F VS+R E C F E L G
Sbjct 1 MIKAILIFNNHGKPRLSKFYQPYSEDTQQQIIRETFHLVSKR-DENVCNFLEGGLLIGGS 59
Query 70 NHRIIYRHFATLFFIFVTDNLESELGLLDLIQVVVQVLDACFENVCELDLVFHYDKINYI 129
++++IYRH+ATL+F+F D+ ESELG+LDLIQV V+ LD CFENVCELDL+FH DK++ I
Sbjct 60 DNKLIYRHYATLYFVFCVDSSESELGILDLIQVFVETLDKCFENVCELDLIFHVDKVHNI 119
Query 130 LDEIIVGGLVMETGVDNIL 148
L E+++GG+V+ET ++ I+
Sbjct 120 LAEMVMGGMVLETNMNEIV 138
> SPAC30D11.05
Length=165
Score = 140 bits (352), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 68/138 (49%), Positives = 93/138 (67%), Gaps = 2/138 (1%)
Query 11 MIKSVVVVNTQGKPRIVRFYDGTPQEKHQHVMKSLFAAVSRRPSEGCCCFTENTDLFGPN 70
MI +V + N +GKPR+ +FY + Q ++ ++AAVS RP C N L
Sbjct 1 MIYAVFIFNNKGKPRLTKFYTPIDESIQQKLIGDIYAAVSTRPPTACNFLESN--LIAGK 58
Query 71 HRIIYRHFATLFFIFVTDNLESELGLLDLIQVVVQVLDACFENVCELDLVFHYDKINYIL 130
+RIIYR +ATL+F+FV D ESELG+LDLIQV V+ LD CF NVCELDLVF + +I+ IL
Sbjct 59 NRIIYRQYATLYFVFVVDEGESELGILDLIQVFVEALDRCFNNVCELDLVFKFQEIHAIL 118
Query 131 DEIIVGGLVMETGVDNIL 148
E++ GGLV+ET ++ I+
Sbjct 119 AEVVSGGLVLETNLNEIV 136
> 7291698
Length=210
Score = 134 bits (337), Expect = 6e-32, Method: Compositional matrix adjust.
Identities = 67/158 (42%), Positives = 101/158 (63%), Gaps = 21/158 (13%)
Query 11 MIKSVVVVNTQGKPRIVRFYDGTPQEKHQHVMKSLFAAVSRRPSEGCCCFTENTDLFG-P 69
MIK+++V N GKPR+ +FY + Q ++K F VS+R + C F E L G
Sbjct 1 MIKAILVFNNHGKPRLSKFYQYFDESLQQQIIKETFQLVSKR-DDNVCNFLEGGSLIGGS 59
Query 70 NHRIIYRHFATLFFIFVTDNLESELGLLDLIQVVVQVLDACFENVCELDLVFHYDKINYI 129
++++IYRH+ATL+F+F D+ ESELG+LDLIQV V+ LD CFENVCELDL+FH D +++I
Sbjct 60 DYKLIYRHYATLYFVFCVDSSESELGILDLIQVFVETLDKCFENVCELDLIFHADAVHHI 119
Query 130 L-------------------DEIIVGGLVMETGVDNIL 148
L E+++GG+V++T +++I+
Sbjct 120 LSGWESYFGNPFRFSYHEFVSELVMGGMVLQTNMNDIM 157
> YJL024c
Length=194
Score = 104 bits (259), Expect = 7e-23, Method: Compositional matrix adjust.
Identities = 55/150 (36%), Positives = 93/150 (62%), Gaps = 12/150 (8%)
Query 11 MIKSVVVVNTQGKPRIVRFYDGTPQEKHQHVMKSLFAAVSRRPSEGCCCF--TENTDLFG 68
MI +V++ N + +PR+V+FY K + +++ ++ +S+R S+ F T + L
Sbjct 1 MIHAVLIFNKKCQPRLVKFYTPVDLPKQKLLLEQVYELISQRNSDFQSSFLVTPPSLLLS 60
Query 69 PNH----------RIIYRHFATLFFIFVTDNLESELGLLDLIQVVVQVLDACFENVCELD 118
+ +IIY+++ATL+F F+ D+ ESEL +LDLIQ V+ LD CF V ELD
Sbjct 61 NENNNDEVNNEDIQIIYKNYATLYFTFIVDDQESELAILDLIQTFVESLDRCFTEVNELD 120
Query 119 LVFHYDKINYILDEIIVGGLVMETGVDNIL 148
L+F++ + +L+EI+ GG+V+ET V+ I+
Sbjct 121 LIFNWQTLESVLEEIVQGGMVIETNVNRIV 150
> At2g19790
Length=143
Score = 95.1 bits (235), Expect = 5e-20, Method: Compositional matrix adjust.
Identities = 46/137 (33%), Positives = 87/137 (63%), Gaps = 6/137 (4%)
Query 12 IKSVVVVNTQGKPRIVRFYDGTPQEKHQHVMKSLFAAVSRRPSEGCCCFTENTDLFGPNH 71
I+ +++VN QG+ R+ ++Y+ E+ + + + R + C F E+ N+
Sbjct 3 IRFILMVNKQGQTRLAQYYEWLTLEERRALEGEIVRKCLARNDQQCS-FVEHR-----NY 56
Query 72 RIIYRHFATLFFIFVTDNLESELGLLDLIQVVVQVLDACFENVCELDLVFHYDKINYILD 131
+I+YR +A+LFF+ D+ E+EL +L+ I ++V+ +D F NVCELD++FH +K +++L+
Sbjct 57 KIVYRRYASLFFMVGVDDDENELAILEFIHLLVETMDKHFGNVCELDIMFHLEKAHFMLE 116
Query 132 EIIVGGLVMETGVDNIL 148
E+++ G ++ET NIL
Sbjct 117 EMVMNGCIVETSKANIL 133
> SPBC685.04c
Length=143
Score = 90.5 bits (223), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 45/138 (32%), Positives = 84/138 (60%), Gaps = 5/138 (3%)
Query 11 MIKSVVVVNTQGKPRIVRFYDGTPQEKHQHVMKSLFAAVSRRPSEGCCCFTENTDLFGPN 70
MI+ +++ N GK R+ ++Y ++ + + +S+R + F E N
Sbjct 1 MIQFILIQNRHGKNRLSKYYVPFDDDEKVRLKARIHQLISQRNQKFQANFLE-----WEN 55
Query 71 HRIIYRHFATLFFIFVTDNLESELGLLDLIQVVVQVLDACFENVCELDLVFHYDKINYIL 130
+++YR +A L+F F D+ +++L +L++I V++LD+ F NVCELDL+F++ K++ IL
Sbjct 56 SKLVYRRYAGLYFCFCVDSTDNDLAILEMIHFFVEILDSFFGNVCELDLIFNFYKVSAIL 115
Query 131 DEIIVGGLVMETGVDNIL 148
DEII+GG + E+ ++L
Sbjct 116 DEIILGGEIGESNKKSVL 133
> Hs4557471
Length=158
Score = 87.8 bits (216), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 45/139 (32%), Positives = 87/139 (62%), Gaps = 8/139 (5%)
Query 11 MIKSVVVVNTQGKPRIVRFYDGTPQEKHQHVMKSLFAAV-SRRPSEGCCCFTENTDLFGP 69
M++ +++ + QGK R+ ++Y T ++ + +++ L V +R+P C F E DL
Sbjct 1 MMRFMLLFSRQGKLRLQKWYLATSDKERKKMVRELMQVVLARKPK--MCSFLEWRDL--- 55
Query 70 NHRIIYRHFATLFFIFVTDNLESELGLLDLIQVVVQVLDACFENVCELDLVFHYDKINYI 129
+++Y+ +A+L+F + ++EL L+LI V++LD F +VCELD++F+++K +I
Sbjct 56 --KVVYKRYASLYFCCAIEGQDNELITLELIHRYVELLDKYFGSVCELDIIFNFEKAYFI 113
Query 130 LDEIIVGGLVMETGVDNIL 148
LDE ++GG V +T ++L
Sbjct 114 LDEFLMGGDVQDTSKKSVL 132
> At4g35410
Length=162
Score = 86.7 bits (213), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 44/137 (32%), Positives = 81/137 (59%), Gaps = 6/137 (4%)
Query 11 MIKSVVVVNTQGKPRIVRFYDGTPQEKHQHVMKSLFAAVSRRPSEGCCCFTENTDLFGPN 70
MI V++V+ QGK R+ ++Y Q++ V++ L + R + C F E
Sbjct 1 MIHFVLLVSRQGKVRLTKWYSPYAQKERSKVIRELSGVILNRGPK-LCNFVE-----WRG 54
Query 71 HRIIYRHFATLFFIFVTDNLESELGLLDLIQVVVQVLDACFENVCELDLVFHYDKINYIL 130
++++Y+ +A+L+F D ++EL +L++I V++LD F +VCELDL+F++ K YIL
Sbjct 55 YKVVYKRYASLYFCMCIDQEDNELEVLEIIHHYVEILDRYFGSVCELDLIFNFHKAYYIL 114
Query 131 DEIIVGGLVMETGVDNI 147
DE+++ G + E+ +
Sbjct 115 DELLIAGELQESSKKTV 131
> CE09797
Length=157
Score = 86.7 bits (213), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 42/139 (30%), Positives = 84/139 (60%), Gaps = 8/139 (5%)
Query 11 MIKSVVVVNTQGKPRIVRFYDGTPQEKHQHVMKSLFAAV-SRRPSEGCCCFTENTDLFGP 69
M++ +++ + QGK R+ ++Y P ++ + + + L + +R+P C F E DL
Sbjct 1 MMQYMLLFSRQGKLRLQKWYTAYPDKQKKKICRELITQILARKPK--MCAFLEYKDL--- 55
Query 70 NHRIIYRHFATLFFIFVTDNLESELGLLDLIQVVVQVLDACFENVCELDLVFHYDKINYI 129
+++Y+ +A+L+F + ++EL L++I V++LD F +VCELD++F+++K +I
Sbjct 56 --KVVYKRYASLYFCCAIEQNDNELITLEVIHRYVELLDKYFGSVCELDIIFNFEKAYFI 113
Query 130 LDEIIVGGLVMETGVDNIL 148
LDE ++ G + ET +L
Sbjct 114 LDEFLLAGEIQETSKKQVL 132
> 7301077
Length=153
Score = 85.9 bits (211), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 43/135 (31%), Positives = 82/135 (60%), Gaps = 8/135 (5%)
Query 15 VVVVNTQGKPRIVRFYDGTPQEKHQHVMKSLFAAV-SRRPSEGCCCFTENTDLFGPNHRI 73
+++ + QGK R+ ++Y P + + + + L + +R+P C F E D +I
Sbjct 1 MLLFSRQGKLRLQKWYMAYPDKVKKKITRELVTTILARKPK--MCSFLEWKDC-----KI 53
Query 74 IYRHFATLFFIFVTDNLESELGLLDLIQVVVQVLDACFENVCELDLVFHYDKINYILDEI 133
+Y+ +A+L+F + ++EL L++I V++LD F +VCELD++F+++K +ILDE+
Sbjct 54 VYKRYASLYFCCAIEQNDNELLTLEIIHRYVELLDKYFGSVCELDIIFNFEKAYFILDEL 113
Query 134 IVGGLVMETGVDNIL 148
++GG + ET N+L
Sbjct 114 LIGGEIQETSKKNVL 128
> Hs22027655
Length=157
Score = 85.5 bits (210), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 45/135 (33%), Positives = 83/135 (61%), Gaps = 8/135 (5%)
Query 15 VVVVNTQGKPRIVRFYDGTPQEKHQHVMKSLFAAV-SRRPSEGCCCFTENTDLFGPNHRI 73
+++ + QGK R+ ++Y ++ + + + L V +R+P C F E DL +I
Sbjct 4 MLLFSRQGKLRLQKWYVPLSDKEKKKITRELVQTVLARKPK--MCSFLEWRDL-----KI 56
Query 74 IYRHFATLFFIFVTDNLESELGLLDLIQVVVQVLDACFENVCELDLVFHYDKINYILDEI 133
+Y+ +A+L+F ++ ++EL L++I V++LD F +VCELD++F+++K +ILDE
Sbjct 57 VYKRYASLYFCCAIEDQDNELITLEIIHRYVELLDKYFGSVCELDIIFNFEKAYFILDEF 116
Query 134 IVGGLVMETGVDNIL 148
++GG V ET N+L
Sbjct 117 LLGGEVQETSKKNVL 131
> Hs22066144
Length=269
Score = 85.5 bits (210), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 45/134 (33%), Positives = 82/134 (61%), Gaps = 8/134 (5%)
Query 16 VVVNTQGKPRIVRFYDGTPQEKHQHVMKSLFAAV-SRRPSEGCCCFTENTDLFGPNHRII 74
+ ++ QGK R+ ++Y ++ + + + L V +R+P C F E DL +I+
Sbjct 114 ISISRQGKLRLQKWYVPLSDKEKKKITRELVQTVLARKPK--MCSFLEWRDL-----KIV 166
Query 75 YRHFATLFFIFVTDNLESELGLLDLIQVVVQVLDACFENVCELDLVFHYDKINYILDEII 134
Y+ +A+L+F ++ ++EL L++I V++LD F +VCELD++F+++K +ILDE +
Sbjct 167 YKRYASLYFCCAIEDQDNELITLEIIHRYVELLDKYFGSVCELDIIFNFEKAYFILDEFL 226
Query 135 VGGLVMETGVDNIL 148
+GG V ET N+L
Sbjct 227 LGGEVQETSKKNVL 240
> Hs16950628
Length=133
Score = 85.1 bits (209), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 44/135 (32%), Positives = 84/135 (62%), Gaps = 8/135 (5%)
Query 11 MIKSVVVVNTQGKPRIVRFYDGTPQEKHQHVMKSLFAAV-SRRPSEGCCCFTENTDLFGP 69
M++ +++ + QGK R+ ++Y T ++ + +++ L V +R+P C F E DL
Sbjct 1 MMRFMLLFSRQGKLRLQKWYLATSDKERKKMVRELMQVVLARKPK--MCSFLEWRDL--- 55
Query 70 NHRIIYRHFATLFFIFVTDNLESELGLLDLIQVVVQVLDACFENVCELDLVFHYDKINYI 129
+++Y+ +A+L+F + ++EL L+LI V++LD F +VCELD++F+++K +I
Sbjct 56 --KVVYKRYASLYFCCAIEGQDNELITLELIHRYVELLDKYFGSVCELDIIFNFEKAYFI 113
Query 130 LDEIIVGGLVMETGV 144
LDE ++GG V +T
Sbjct 114 LDEFLMGGDVQDTST 128
> At2g17380
Length=161
Score = 84.7 bits (208), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 42/133 (31%), Positives = 79/133 (59%), Gaps = 6/133 (4%)
Query 15 VVVVNTQGKPRIVRFYDGTPQEKHQHVMKSLFAAVSRRPSEGCCCFTENTDLFGPNHRII 74
V++V+ QGK R+ ++Y Q++ V++ L + R + C F E ++++
Sbjct 5 VLLVSRQGKVRLTKWYSPYTQKERSKVIRELSGVILNRGPK-LCNFIE-----WRGYKVV 58
Query 75 YRHFATLFFIFVTDNLESELGLLDLIQVVVQVLDACFENVCELDLVFHYDKINYILDEII 134
Y+ +A+L+F D ++EL +L++I V++LD F +VCELDL+F++ K YILDE++
Sbjct 59 YKRYASLYFCMCIDEADNELEVLEIIHHYVEILDRYFGSVCELDLIFNFHKAYYILDELL 118
Query 135 VGGLVMETGVDNI 147
+ G + E+ +
Sbjct 119 IAGELQESSKKTV 131
> At1g47830
Length=142
Score = 84.0 bits (206), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 49/138 (35%), Positives = 77/138 (55%), Gaps = 6/138 (4%)
Query 11 MIKSVVVVNTQGKPRIVRFYDGTPQEKHQHVMKSLFAAVSRRPSEGCCCFTENTDLFGPN 70
MI+ +++ N QGK R+ ++Y + + V + V R ++ FT +
Sbjct 1 MIRFILLQNRQGKTRLAKYYVPLEESEKHKVEYEVHRLVVNRDAK----FTNFVEF--RT 54
Query 71 HRIIYRHFATLFFIFVTDNLESELGLLDLIQVVVQVLDACFENVCELDLVFHYDKINYIL 130
H++IYR +A LFF D ++EL L+ I + V++LD F NVCELDLVF++ K+ IL
Sbjct 55 HKVIYRRYAGLFFSVCVDITDNELAYLESIHLFVEILDHFFSNVCELDLVFNFHKVYLIL 114
Query 131 DEIIVGGLVMETGVDNIL 148
DE I+ G + ET I+
Sbjct 115 DEFILAGELQETSKRAII 132
> HsM5901934
Length=144
Score = 83.2 bits (204), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 46/138 (33%), Positives = 78/138 (56%), Gaps = 6/138 (4%)
Query 11 MIKSVVVVNTQGKPRIVRFYDGTPQEKHQHVMKSLFAAVSRRPSEGCCCFTENTDLFGPN 70
MIK ++VN QG+ R+ ++Y+ K + + + R +E C F E D
Sbjct 1 MIKFFLMVNKQGQTRLSKYYEHVDINKRTLLETEVIKSCLSRSNEQCS-FIEYKDF---- 55
Query 71 HRIIYRHFATLFFIFVTDNLESELGLLDLIQVVVQVLDACFENVCELDLVFHYDKINYIL 130
++IYR +A LF + ++ E+E+ + + I V+VLD F V ELD++F+ DK++ IL
Sbjct 56 -KLIYRQYAALFIVVGVNDTENEMAIYEFIHNFVEVLDEYFSRVSELDIMFNLDKVHIIL 114
Query 131 DEIIVGGLVMETGVDNIL 148
DE+++ G ++ET IL
Sbjct 115 DEMVLNGCIVETNRARIL 132
> YJR058c
Length=147
Score = 83.2 bits (204), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 47/141 (33%), Positives = 81/141 (57%), Gaps = 6/141 (4%)
Query 10 IMIKSVVVVNTQGKPRIVRFYD--GTPQEKHQHVMKSLFAAVSRRPSEGCCCFTENTDLF 67
+ ++ ++ N QG R+VR++D + ++ Q + ++ +S R + F E F
Sbjct 1 MAVQFILCFNKQGVVRLVRWFDVHSSDPQRSQDAIAQIYRLISSRDHKHQSNFVE----F 56
Query 68 GPNHRIIYRHFATLFFIFVTDNLESELGLLDLIQVVVQVLDACFENVCELDLVFHYDKIN 127
+ ++IYR +A L+F+ D L+ E L I + V+VLDA F NVCELD+VF++ K+
Sbjct 57 SDSTKLIYRRYAGLYFVMGVDLLDDEPIYLCHIHLFVEVLDAFFGNVCELDIVFNFYKVY 116
Query 128 YILDEIIVGGLVMETGVDNIL 148
I+DE+ +GG + E D +L
Sbjct 117 MIMDEMFIGGEIQEISKDMLL 137
> SPAP27G11.06c
Length=162
Score = 82.8 bits (203), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 43/138 (31%), Positives = 85/138 (61%), Gaps = 8/138 (5%)
Query 12 IKSVVVVNTQGKPRIVRFYDGTPQEKHQHVMKSLFAAV-SRRPSEGCCCFTENTDLFGPN 70
IK ++V+ QGK R+ ++++ ++ +++ + + V +R+P C F E
Sbjct 3 IKFFLLVSRQGKVRLAKWFNTLSIKERAKIIRDVSSLVITRKPK--MCNFVEY-----KG 55
Query 71 HRIIYRHFATLFFIFVTDNLESELGLLDLIQVVVQVLDACFENVCELDLVFHYDKINYIL 130
+I+YR +A+LFF+ + ++EL +L++I V+ LD F NVCELDL+F+++K Y++
Sbjct 56 EKIVYRRYASLFFVCGIEQDDNELIILEVIHKFVECLDKYFGNVCELDLIFNFEKAYYVM 115
Query 131 DEIIVGGLVMETGVDNIL 148
+E+++ G + E+ N+L
Sbjct 116 EELLLAGELQESSKTNVL 133
> Hs18551999
Length=164
Score = 79.7 bits (195), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 40/133 (30%), Positives = 79/133 (59%), Gaps = 6/133 (4%)
Query 11 MIKSVVVVNTQGKPRIVRFYDGTPQEKHQHVMKSLFAAVSRRPSEGCCCFTENTDLFGPN 70
MI +++ + QGK R+ ++Y P ++ + + + + + R F + +L
Sbjct 1 MIHFILLFSRQGKLRLQKWYITLPDKERKKITREIVQIILSR-GHRTSSFVDWKEL---- 55
Query 71 HRIIYRHFATLFFIFVTDNLESELGLLDLIQVVVQVLDACFENVCELDLVFHYDKINYIL 130
+++Y+ +A+L+F +N ++EL L+++ V++LD F NVCELD++F+++K +IL
Sbjct 56 -KLVYKRYASLYFCCAIENQDNELLTLEIVHRYVELLDKYFGNVCELDIIFNFEKAYFIL 114
Query 131 DEIIVGGLVMETG 143
DE I+GG + ET
Sbjct 115 DEFIIGGEIQETS 127
> Hs4757996
Length=142
Score = 77.0 bits (188), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 45/138 (32%), Positives = 79/138 (57%), Gaps = 6/138 (4%)
Query 11 MIKSVVVVNTQGKPRIVRFYDGTPQEKHQHVMKSLFAAVSRRPSEGCCCFTENTDLFGPN 70
MI+ +++ N GK R+ ++Y ++ Q +++ + A V+ R ++ F E N
Sbjct 1 MIRFILIQNRAGKTRLAKWYMQFDDDEKQKLIEEVHAVVTVRDAKHTN-FVE-----FRN 54
Query 71 HRIIYRHFATLFFIFVTDNLESELGLLDLIQVVVQVLDACFENVCELDLVFHYDKINYIL 130
+IIYR +A L+F D +++L L+ I V+VL+ F NVCELDLVF++ K+ ++
Sbjct 55 FKIIYRRYAGLYFCICVDVNDNKLAYLEGIHNFVEVLNEYFHNVCELDLVFNFYKVYTVV 114
Query 131 DEIIVGGLVMETGVDNIL 148
DE+ + G + ET +L
Sbjct 115 DEMFLAGEIRETSQTKVL 132
> 7300728
Length=142
Score = 76.3 bits (186), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 44/138 (31%), Positives = 78/138 (56%), Gaps = 6/138 (4%)
Query 11 MIKSVVVVNTQGKPRIVRFYDGTPQEKHQHVMKSLFAAVSRRPSEGCCCFTENTDLFGPN 70
MI+ +++ N GK R+ ++Y ++ Q +++ + A V+ R ++ F E N
Sbjct 1 MIRFILIQNRAGKTRLAKWYMNFDDDEKQKLIEEVHAVVTVRDAKHTN-FVE-----FRN 54
Query 71 HRIIYRHFATLFFIFVTDNLESELGLLDLIQVVVQVLDACFENVCELDLVFHYDKINYIL 130
+I+YR +A L+F D ++ L L+ I V+VL+ F NVCELDLVF++ K+ ++
Sbjct 55 FKIVYRRYAGLYFCICVDVNDNNLCYLEAIHNFVEVLNEYFHNVCELDLVFNFYKVYSVV 114
Query 131 DEIIVGGLVMETGVDNIL 148
DE+ + G + ET +L
Sbjct 115 DEMFLAGEIRETSQTKVL 132
> CE07018
Length=142
Score = 76.3 bits (186), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 44/138 (31%), Positives = 78/138 (56%), Gaps = 6/138 (4%)
Query 11 MIKSVVVVNTQGKPRIVRFYDGTPQEKHQHVMKSLFAAVSRRPSEGCCCFTENTDLFGPN 70
MI+ +++ N GK R+ ++Y ++ Q +++ + A V+ R ++ F E N
Sbjct 1 MIRFILIQNRAGKTRLAKWYMHFDDDEKQKLIEEVHACVTVRDAKHTN-FVE-----FRN 54
Query 71 HRIIYRHFATLFFIFVTDNLESELGLLDLIQVVVQVLDACFENVCELDLVFHYDKINYIL 130
+I+YR +A L+F D ++ L L+ I V+VL+ F NVCELDLVF++ K+ ++
Sbjct 55 FKIVYRRYAGLYFCICVDITDNNLYYLEAIHNFVEVLNEYFHNVCELDLVFNFYKVYTVV 114
Query 131 DEIIVGGLVMETGVDNIL 148
DE+ + G + ET +L
Sbjct 115 DEMFLAGEIRETSQTKVL 132
> YLR170c
Length=156
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 41/139 (29%), Positives = 82/139 (58%), Gaps = 8/139 (5%)
Query 12 IKSVVVVNTQGKPRIVRFYDGTPQEKHQHVMKSLFAAVSRRPSEGCCCFTENTDLFGPNH 71
+K +++V+ QGK R+ ++Y + ++K L + R + C N +H
Sbjct 4 LKYLLLVSRQGKIRLKKWYTAMSAGEKAKIVKDLTPTILARKPKMCNIIEYN------DH 57
Query 72 RIIYRHFATLFFIF-VTDNLESELGLLDLIQVVVQVLDACFENVCELDLVFHYDKINYIL 130
+++Y+ +A+L+FI +T ++++EL L++I V+ +D F NVCELD++F++ K+ IL
Sbjct 58 KVVYKRYASLYFIVGMTPDVDNELLTLEIIHRFVETMDTYFGNVCELDIIFNFSKVYDIL 117
Query 131 DEIIV-GGLVMETGVDNIL 148
+E+I+ G + E+ +L
Sbjct 118 NEMIMCDGSIAESSRKEVL 136
> Hs21361394
Length=159
Score = 60.1 bits (144), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 35/111 (31%), Positives = 58/111 (52%), Gaps = 6/111 (5%)
Query 11 MIKSVVVVNTQGKPRIVRFYDGTPQEKHQHVMKSLFAAVSRRPSEGCCCFTENTDLFGPN 70
MIK ++VN QG+ R+ ++Y+ K + + + R +E C F E D
Sbjct 1 MIKFFLMVNKQGQTRLSKYYEHVDINKRTLLETEVIKSCLSRSNEQCS-FIEYKDF---- 55
Query 71 HRIIYRHFATLFFIFVTDNLESELGLLDLIQVVVQVLDACFENVCELDLVF 121
++IYR +A LF + ++ E+E+ + + I V+VLD F V ELD+ F
Sbjct 56 -KLIYRQYAALFIVVGVNDTENEMAIYEFIHNFVEVLDEYFSRVSELDVSF 105
> Hs7705983
Length=210
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 43/142 (30%), Positives = 76/142 (53%), Gaps = 13/142 (9%)
Query 5 IQFPSI-MIKSVVVVNTQGKPRIVRFYDGT-PQEKHQHVM-KSLFAAVSRRPSEGCCCFT 61
+Q PS+ IK+V +++ G+ + ++YD T P K Q V K++F SR SE
Sbjct 38 LQEPSLYTIKAVFILDNDGRRLLAKYYDDTFPSMKEQMVFEKNVFNKTSRTESEIA---- 93
Query 62 ENTDLFGPNHRIIYRHFATLFFIFVTDNLESELGLLDLIQVVVQVLDACF-ENVCELDLV 120
FG I+Y++ LF V + E+EL L+ ++ + + L+ +NV + L+
Sbjct 94 ----FFG-GMTIVYKNSIDLFLYVVGSSYENELMLMSVLTCLFESLNHMLRKNVEKRWLL 148
Query 121 FHYDKINYILDEIIVGGLVMET 142
+ D +LDEI+ GG+++E+
Sbjct 149 ENMDGAFLVLDEIVDGGVILES 170
> Hs11038643
Length=104
Score = 45.1 bits (105), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 31/138 (22%), Positives = 58/138 (42%), Gaps = 44/138 (31%)
Query 11 MIKSVVVVNTQGKPRIVRFYDGTPQEKHQHVMKSLFAAVSRRPSEGCCCFTENTDLFGPN 70
MI+ +++ N GK R+ ++Y ++ Q +++ + A V+ R ++
Sbjct 1 MIRFILIQNRAGKTRLAKWYMQFDDDEKQKLIEEVHAVVTVRDAKHTN------------ 48
Query 71 HRIIYRHFATLFFIFVTDNLESELGLLDLIQVVVQVLDACFENVCELDLVFHYDKINYIL 130
V+VL+ F NVCELDLVF++ K+ ++
Sbjct 49 --------------------------------FVEVLNEYFHNVCELDLVFNFYKVYTVV 76
Query 131 DEIIVGGLVMETGVDNIL 148
DE+ + G + ET +L
Sbjct 77 DEMFLAGEIRETSQTKVL 94
> ECU08g0150
Length=98
Score = 45.1 bits (105), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 22/70 (31%), Positives = 40/70 (57%), Gaps = 1/70 (1%)
Query 73 IIYRHFATLFFIFVTDNLESELGLLDLIQVVVQVLDACFENVCELDLVFHYDKINYILDE 132
+++ F +F FV +N E+E+ +L LI ++ + D F VCEL ++++ + + ILD
Sbjct 17 VVFNRFGNIFMAFVVEN-ENEMYILSLINNLMSIYDRFFTKVCELHFIYNFKETHIILDN 75
Query 133 IIVGGLVMET 142
I G +E
Sbjct 76 YIANGKCIEN 85
> CE11494
Length=184
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 42/148 (28%), Positives = 66/148 (44%), Gaps = 38/148 (25%)
Query 12 IKSVVVVNTQGKPRIVRFYD----GTPQEKHQHVMKSLFAAVSRRPS------EGCCCFT 61
IK +V+++ G + ++YD GT +E + KSLF+ SR S +G C
Sbjct 14 IKGIVILDQDGNRVLAKYYDRTTFGTVKE-QKAFEKSLFSKTSRNTSADILLLDGVTC-- 70
Query 62 ENTDLFGPNHRIIYRHFATLFFIFVTDNLESEL-------GLLDLIQVVVQVLDACFENV 114
+YR L+F + E+EL L D + VV++ +NV
Sbjct 71 ------------LYRSNVDLYFYVLGSTRENELFLDATLTCLYDAVSVVLR------KNV 112
Query 115 CELDLVFHYDKINYILDEIIVGGLVMET 142
+ L+ D I I+DEI G++MET
Sbjct 113 EKKALIDSMDTIMLIIDEICDEGIIMET 140
> Hs20539280
Length=185
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 18/46 (39%), Positives = 33/46 (71%), Gaps = 0/46 (0%)
Query 92 SELGLLDLIQVVVQVLDACFENVCELDLVFHYDKINYILDEIIVGG 137
+EL L+ I +++L+ F +VCELD+VF+++++ +ILDE + G
Sbjct 120 NELTALETIHHYMELLNKYFGSVCELDVVFNFEEVYFILDEFFLEG 165
> At1g15370
Length=147
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 39/141 (27%), Positives = 61/141 (43%), Gaps = 10/141 (7%)
Query 11 MIKSVVVVNTQGKPRIVRFYDGTPQEKHQHVMKSLFAAVSRRPSEGCCCFTENTDLFGPN 70
MI +V+ N+ G I RF +G P E+ H +S + +G +N +L
Sbjct 1 MILAVLFANSVGNVLIERF-NGVPAEERLH-WRSFLVKLGADNLKG----VKNEELLVAC 54
Query 71 HR---IIYRHFATLFFIFVTDNLESELGLLDLIQVV-VQVLDACFENVCELDLVFHYDKI 126
H+ I+Y + V + EL L + I ++ V D C + E + Y +I
Sbjct 55 HKSVYIVYTMLGDVSIFLVGKDEYDELALAETIYIITAAVKDVCGKPPTERVFLDKYGRI 114
Query 127 NYILDEIIVGGLVMETGVDNI 147
LDEI+ GL+ T D I
Sbjct 115 CLCLDEIVWNGLLENTDKDRI 135
> Hs7706337
Length=177
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 32/139 (23%), Positives = 67/139 (48%), Gaps = 13/139 (9%)
Query 8 PSI-MIKSVVVVNTQGKPRIVRFYDGT-PQEKHQHVM-KSLFAAVSRRPSEGCCCFTENT 64
PS+ +K++++++ G ++YD T P K Q K++F R SE
Sbjct 8 PSLYTVKAILILDNDGDRLFAKYYDDTYPSVKEQKAFEKNIFNKTHRTDSEIALL----- 62
Query 65 DLFGPNHRIIYRHFATLFFIFVTDNLESELGLLDLIQVVVQVLDACF-ENVCELDLVFHY 123
++Y+ L+F + + E+EL L+ ++ + L +NV + L+ +
Sbjct 63 ----EGLTVVYKSSIDLYFYVIGSSYENELMLMAVLNCLFDSLSQMLRKNVEKRALLENM 118
Query 124 DKINYILDEIIVGGLVMET 142
+ + +DEI+ GG+++E+
Sbjct 119 EGLFLAVDEIVDGGVILES 137
> At1g60970
Length=162
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 38/143 (26%), Positives = 66/143 (46%), Gaps = 20/143 (13%)
Query 12 IKSVVVVNTQGKPRIVRFY-DGTPQEKHQHVM-KSLFAAVSRRPSEGCCCFTENTDLFGP 69
+K++++++++GK V++Y D P Q KS+F + E T L
Sbjct 7 VKNILLLDSEGKRVAVKYYSDDWPTNSAQEAFEKSVFTKTQKT---NARTEVEVTAL--E 61
Query 70 NHRIIYRHFATLFFIFVTDNLESEL-------GLLDLIQVVVQVLDACFENVCELDLVFH 122
N+ ++Y+ L F E+EL GL D + ++++ E + LDL+F
Sbjct 62 NNIVVYKFVQDLHFFVTGGEEENELILASVLEGLFDAVTLLLRSNVDKREALDNLDLIF- 120
Query 123 YDKINYILDEIIVGGLVMETGVD 145
DEII GG+V+ET +
Sbjct 121 -----LSFDEIIDGGIVLETDAN 138
> At3g09800
Length=180
Score = 32.7 bits (73), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 32/135 (23%), Positives = 66/135 (48%), Gaps = 8/135 (5%)
Query 11 MIKSVVVVNTQGKPRIVRFY-DGTPQEKHQ-HVMKSLFAAVSRRPSEGCCCFTENTDLFG 68
++K++++++++GK V++Y D P + K +F+ S+ + TE
Sbjct 8 LVKNILLLDSEGKRVAVKYYSDDWPTNAAKLSFEKYVFSKTSKTNAR-----TEAEITLL 62
Query 69 PNHRIIYRHFATLFFIFVTDNLESELGLLDLIQVVVQVLDACF-ENVCELDLVFHYDKIN 127
++ I+Y+ L F E+EL L ++Q + NV +++ + + D I
Sbjct 63 DSNIIVYKFAQDLHFFVTGGENENELILASVLQGFFDAVALLLRSNVEKMEALENLDLIF 122
Query 128 YILDEIIVGGLVMET 142
LDE++ G+V+ET
Sbjct 123 LCLDEMVDQGVVLET 137
> 7303942
Length=660
Score = 31.6 bits (70), Expect = 0.60, Method: Composition-based stats.
Identities = 16/44 (36%), Positives = 23/44 (52%), Gaps = 1/44 (2%)
Query 73 IIYRHFATLFFIFVTDNLESELGLLDLIQVVVQVLDACFENVCE 116
I Y+HF +L +F + E L D++ V QV C+ VCE
Sbjct 33 IQYQHFLSLLEVFALNPSEENKSLDDIVMFVAQVAQ-CYPAVCE 75
> At2g03730
Length=446
Score = 31.2 bits (69), Expect = 0.69, Method: Compositional matrix adjust.
Identities = 20/61 (32%), Positives = 30/61 (49%), Gaps = 6/61 (9%)
Query 27 VRFYDGTP---QEKHQHVMKSLFAAVSRRPSEGC---CCFTENTDLFGPNHRIIYRHFAT 80
+R DG+P + + Q V+K L AA+ RR SEG C ++ L RI + T
Sbjct 305 IRHTDGSPVKSEAERQRVIKCLKAAIQRRVSEGLKLELCTSDRVGLLSDVTRIFRENSLT 364
Query 81 L 81
+
Sbjct 365 V 365
> At1g69040
Length=451
Score = 30.4 bits (67), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 21/61 (34%), Positives = 30/61 (49%), Gaps = 6/61 (9%)
Query 27 VRFYDGTP---QEKHQHVMKSLFAAVSRRPSEGC---CCFTENTDLFGPNHRIIYRHFAT 80
VR DG+P + + Q V++ L AA+ RR SEG C T+ L RI + T
Sbjct 303 VRHIDGSPVKSEAEKQRVIQCLEAAIKRRVSEGLKLELCTTDRVGLLSNVTRIFRENSLT 362
Query 81 L 81
+
Sbjct 363 V 363
> SPCC285.08
Length=240
Score = 30.0 bits (66), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 30/129 (23%), Positives = 64/129 (49%), Gaps = 10/129 (7%)
Query 10 IMIKSVVVVNTQGKPRIVRFYDGTPQEKHQHVMKSLFAAVSRRPSEGCCCFTENTDLFGP 69
+++ +V +VN GK I R + + + + ++ S A VS + ++NT +
Sbjct 1 MVVLAVSIVNRGGKAIISRQFREMSRVRVESLLSSFPALVSEK--------SQNTTVESD 52
Query 70 NHRIIYRHFATLFFIFVTDNLESE-LGLLDLIQVVVQVLDACFENVCELDLVFHYDKINY 128
N R +Y+ L+ + +T NL+S L +D + ++ QV+ + ++ E +++ + +I
Sbjct 53 NVRFVYQPLDELYIVLIT-NLQSNILQDIDTLHLLSQVVTSICSSLEEREILEYAFEIFT 111
Query 129 ILDEIIVGG 137
DE G
Sbjct 112 AFDEATSLG 120
> YPL010w
Length=189
Score = 30.0 bits (66), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 28/142 (19%), Positives = 65/142 (45%), Gaps = 8/142 (5%)
Query 12 IKSVVVVNTQGKPRIVRFYDGTPQEKHQHVMKSLFAAVSRRPSEGCCCF--TENTD---L 66
+++V++++ QG+ ++Y P + + LF +V ++ + T D L
Sbjct 9 VQAVLILDQQGERIYAKYY--QPPHRSDEGHQLLFNSVKKQKEFEKQLYRKTHKQDSEIL 66
Query 67 FGPNHRIIYRHFATLFFIFVTDNLESELGLLDLIQVVVQVLDACFEN-VCELDLVFHYDK 125
+H ++Y+ + + V E+E+ L + LD + + + ++ +YD
Sbjct 67 IFEDHLVLYKEYIDITIYLVASLEENEIVLQQGFSAIRGALDLILNSGMDKKNIQENYDM 126
Query 126 INYILDEIIVGGLVMETGVDNI 147
+ +DE I G+++ET + I
Sbjct 127 VLLAIDETIDNGVILETDSNTI 148
> At5g06450
Length=206
Score = 28.9 bits (63), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 18/70 (25%), Positives = 31/70 (44%), Gaps = 1/70 (1%)
Query 29 FYD-GTPQEKHQHVMKSLFAAVSRRPSEGCCCFTENTDLFGPNHRIIYRHFATLFFIFVT 87
++D G P+ + + + + + C F F N + +YR FA+ F FV
Sbjct 54 YWDVGFPETETKTKTSGWSLSSVKLSTRNLCLFLRLPKPFHDNLKDLYRFFASKFVTFVG 113
Query 88 DNLESELGLL 97
+E +L LL
Sbjct 114 VQIEEDLDLL 123
> At1g48860
Length=521
Score = 28.5 bits (62), Expect = 4.5, Method: Composition-based stats.
Identities = 15/43 (34%), Positives = 25/43 (58%), Gaps = 0/43 (0%)
Query 104 VQVLDACFENVCELDLVFHYDKINYILDEIIVGGLVMETGVDN 146
+ +L A E +D + + D INY+LD + + GL +ET +N
Sbjct 106 ILLLAALSEGTTVVDNLLNSDDINYMLDALKILGLNVETHSEN 148
> SPAC644.06c
Length=593
Score = 28.1 bits (61), Expect = 6.1, Method: Composition-based stats.
Identities = 28/122 (22%), Positives = 51/122 (41%), Gaps = 13/122 (10%)
Query 5 IQFPSIMIKSVVVVNTQGKPRIVRFYDGTPQEKHQHVM------KSLFAAVSRR----PS 54
I++ SI ++ ++++ P I+R YD +H ++ LF + +
Sbjct 45 IRYASIGME-ILMMRLLRHPNILRLYDVWTDHQHMYLALEYVPDGELFHYIRKHGPLSER 103
Query 55 EGCCCFTENTDLFGPNHRIIYRHF-ATLFFIFVTDNLESELGLLDLIQVVVQVLDACFEN 113
E ++ D HR +RH L I + N E ++ + D V+ D+C EN
Sbjct 104 EAAHYLSQILDAVAHCHRFRFRHRDLKLENILIKVN-EQQIKIADFGMATVEPNDSCLEN 162
Query 114 VC 115
C
Sbjct 163 YC 164
> YGR217w
Length=2039
Score = 27.7 bits (60), Expect = 7.9, Method: Compositional matrix adjust.
Identities = 14/58 (24%), Positives = 29/58 (50%), Gaps = 2/58 (3%)
Query 29 FYDGTPQEKHQHVMKSLFAAVSRRPSEGCCCFTENTDLFGPNHRIIYRHFATLFFIFV 86
+Y +PQ + + + L + + ++G F ++TDL+ N R + H F+F+
Sbjct 1156 YYFFSPQHRFRRFCQRLVPPSTGKRTDGSRFFEDSTDLY--NKRSYFHHIERDVFVFI 1211
Lambda K H
0.328 0.145 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1821716300
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40