bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0319_orf1
Length=118
Score E
Sequences producing significant alignments: (Bits) Value
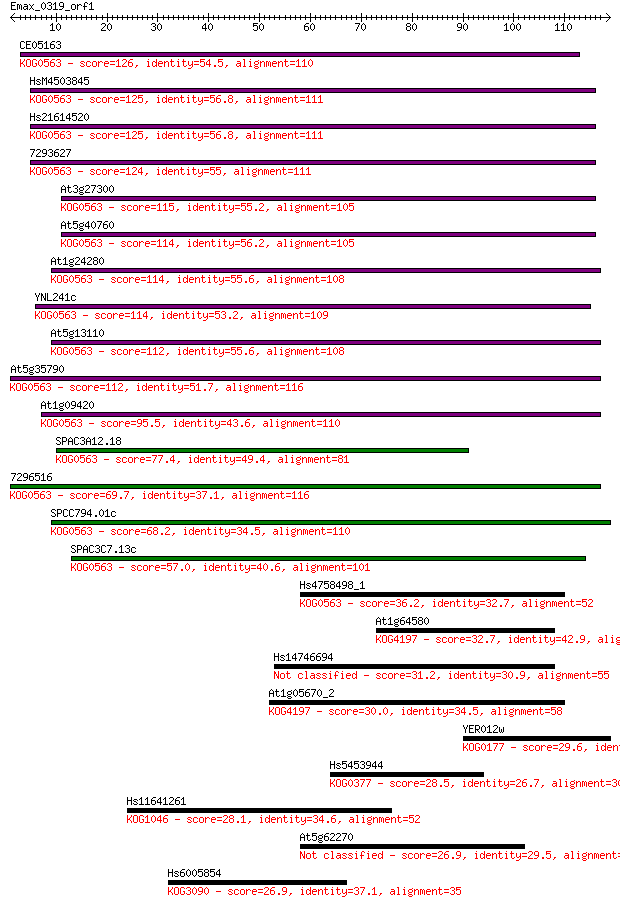
CE05163 126 1e-29
HsM4503845 125 2e-29
Hs21614520 125 2e-29
7293627 124 6e-29
At3g27300 115 3e-26
At5g40760 114 3e-26
At1g24280 114 4e-26
YNL241c 114 6e-26
At5g13110 112 1e-25
At5g35790 112 2e-25
At1g09420 95.5 2e-20
SPAC3A12.18 77.4 6e-15
7296516 69.7 1e-12
SPCC794.01c 68.2 4e-12
SPAC3C7.13c 57.0 8e-09
Hs4758498_1 36.2 0.015
At1g64580 32.7 0.15
Hs14746694 31.2 0.49
At1g05670_2 30.0 0.98
YER012w 29.6 1.3
Hs5453944 28.5 3.1
Hs11641261 28.1 3.6
At5g62270 26.9 8.5
Hs6005854 26.9 9.3
> CE05163
Length=522
Score = 126 bits (316), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 60/110 (54%), Positives = 79/110 (71%), Gaps = 0/110 (0%)
Query 3 YPEEELSENELVIIVQPREAIYLKFYTKKPGLASGLQLTELDLSVMDRLQVDRLPDAYER 62
YP EL +ELV+ VQP EA+Y+K TKKPG+ G++ TELDL+ +R + RLPDAYER
Sbjct 388 YPSGELKRSELVMRVQPNEAVYMKLMTKKPGMGFGVEETELDLTYNNRFKEVRLPDAYER 447
Query 63 LLLDVIKGDRQNFVRTDELREAWRIFTPLLKQIDEPGVKPEPYPYGSHGP 112
L L+V G + NFVRTDEL AWRI TP+L+++ + V+P Y +GS GP
Sbjct 448 LFLEVFMGSQINFVRTDELEYAWRILTPVLEELKKKKVQPVQYKFGSRGP 497
> HsM4503845
Length=515
Score = 125 bits (313), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 63/111 (56%), Positives = 78/111 (70%), Gaps = 0/111 (0%)
Query 5 EEELSENELVIIVQPREAIYLKFYTKKPGLASGLQLTELDLSVMDRLQVDRLPDAYERLL 64
++ NELVI VQP EA+Y K TKKPG+ + +ELDL+ +R + +LPDAYERL+
Sbjct 382 HQQCKRNELVIRVQPNEAVYTKMMTKKPGMFFNPEESELDLTYGNRYKNVKLPDAYERLI 441
Query 65 LDVIKGDRQNFVRTDELREAWRIFTPLLKQIDEPGVKPEPYPYGSHGPESA 115
LDV G + +FVR+DELREAWRIFTPLL QI+ KP PY YGS GP A
Sbjct 442 LDVFCGSQMHFVRSDELREAWRIFTPLLHQIELEKPKPIPYIYGSRGPTEA 492
> Hs21614520
Length=515
Score = 125 bits (313), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 63/111 (56%), Positives = 78/111 (70%), Gaps = 0/111 (0%)
Query 5 EEELSENELVIIVQPREAIYLKFYTKKPGLASGLQLTELDLSVMDRLQVDRLPDAYERLL 64
++ NELVI VQP EA+Y K TKKPG+ + +ELDL+ +R + +LPDAYERL+
Sbjct 382 HQQCKRNELVIRVQPNEAVYTKMMTKKPGMFFNPEESELDLTYGNRYKNVKLPDAYERLI 441
Query 65 LDVIKGDRQNFVRTDELREAWRIFTPLLKQIDEPGVKPEPYPYGSHGPESA 115
LDV G + +FVR+DELREAWRIFTPLL QI+ KP PY YGS GP A
Sbjct 442 LDVFCGSQMHFVRSDELREAWRIFTPLLHQIELEKPKPIPYIYGSRGPTEA 492
> 7293627
Length=524
Score = 124 bits (310), Expect = 6e-29, Method: Compositional matrix adjust.
Identities = 61/111 (54%), Positives = 77/111 (69%), Gaps = 0/111 (0%)
Query 5 EEELSENELVIIVQPREAIYLKFYTKKPGLASGLQLTELDLSVMDRLQVDRLPDAYERLL 64
E NELVI VQP EA+Y K TK PG+ ++ TELDL+ R + LPDAYERL+
Sbjct 387 EGNTKRNELVIRVQPGEALYFKMMTKSPGITFDIEETELDLTYEHRYKDSYLPDAYERLI 446
Query 65 LDVIKGDRQNFVRTDELREAWRIFTPLLKQIDEPGVKPEPYPYGSHGPESA 115
LDV G + +FVR+DELREAWRIFTP+L QI++ ++P Y YGS GP+ A
Sbjct 447 LDVFCGSQMHFVRSDELREAWRIFTPILHQIEKEHIRPITYQYGSRGPKEA 497
> At3g27300
Length=516
Score = 115 bits (287), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 58/105 (55%), Positives = 72/105 (68%), Gaps = 0/105 (0%)
Query 11 NELVIIVQPREAIYLKFYTKKPGLASGLQLTELDLSVMDRLQVDRLPDAYERLLLDVIKG 70
NE VI +QP EA+Y+K K+PGL +ELDLS R Q +P+AYERL+LD I+G
Sbjct 390 NEFVIRLQPSEAMYMKLTVKQPGLEMQTVQSELDLSYKQRYQDVSIPEAYERLILDTIRG 449
Query 71 DRQNFVRTDELREAWRIFTPLLKQIDEPGVKPEPYPYGSHGPESA 115
D+Q+FVR DEL+ AW IFTPLL +ID+ VK PY GS GP A
Sbjct 450 DQQHFVRRDELKAAWEIFTPLLHRIDKGEVKSVPYKQGSRGPAEA 494
> At5g40760
Length=515
Score = 114 bits (286), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 59/105 (56%), Positives = 73/105 (69%), Gaps = 0/105 (0%)
Query 11 NELVIIVQPREAIYLKFYTKKPGLASGLQLTELDLSVMDRLQVDRLPDAYERLLLDVIKG 70
NE VI +QP EA+Y+K K+PGL +ELDLS R Q +P+AYERL+LD IKG
Sbjct 389 NEFVIRLQPSEAMYMKLTVKQPGLDMNTVQSELDLSYGQRYQGVAIPEAYERLILDTIKG 448
Query 71 DRQNFVRTDELREAWRIFTPLLKQIDEPGVKPEPYPYGSHGPESA 115
D+Q+FVR DEL+ AW IFTPLL +ID+ VK PY GS GP+ A
Sbjct 449 DQQHFVRRDELKVAWEIFTPLLHRIDKGEVKSIPYKPGSRGPKEA 493
> At1g24280
Length=599
Score = 114 bits (285), Expect = 4e-26, Method: Compositional matrix adjust.
Identities = 60/108 (55%), Positives = 74/108 (68%), Gaps = 1/108 (0%)
Query 9 SENELVIIVQPREAIYLKFYTKKPGLASGLQLTELDLSVMDRLQVDRLPDAYERLLLDVI 68
+ NELVI VQP EAIYLK K PGL L + L+L R + +PDAYERLLLD I
Sbjct 476 TTNELVIRVQPDEAIYLKINNKVPGLGMRLDQSNLNLLYSARYSKE-IPDAYERLLLDAI 534
Query 69 KGDRQNFVRTDELREAWRIFTPLLKQIDEPGVKPEPYPYGSHGPESAY 116
+G+R+ F+R+DEL AW +FTPLLK+I+E PE YPYGS GP A+
Sbjct 535 EGERRLFIRSDELDAAWALFTPLLKEIEEKKTTPEFYPYGSRGPVGAH 582
> YNL241c
Length=505
Score = 114 bits (284), Expect = 6e-26, Method: Compositional matrix adjust.
Identities = 58/110 (52%), Positives = 75/110 (68%), Gaps = 1/110 (0%)
Query 6 EELSENELVIIVQPREAIYLKFYTKKPGLASGLQLTELDLSVMDRLQVDRLPDAYERLLL 65
+++ NELVI VQP A+YLKF K PGL++ Q+T+L+L+ R Q +P+AYE L+
Sbjct 365 KDIPNNELVIRVQPDAAVYLKFNAKTPGLSNATQVTDLNLTYASRYQDFWIPEAYEVLIR 424
Query 66 DVIKGDRQNFVRTDELREAWRIFTPLLKQIDEP-GVKPEPYPYGSHGPES 114
D + GD NFVR DEL +W IFTPLLK I+ P G PE YPYGS GP+
Sbjct 425 DALLGDHSNFVRDDELDISWGIFTPLLKHIERPDGPTPEIYPYGSRGPKG 474
> At5g13110
Length=593
Score = 112 bits (281), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 60/108 (55%), Positives = 74/108 (68%), Gaps = 1/108 (0%)
Query 9 SENELVIIVQPREAIYLKFYTKKPGLASGLQLTELDLSVMDRLQVDRLPDAYERLLLDVI 68
+ NELVI VQP EAIYLK K PGL L + L+L R + +PDAYERLLLD I
Sbjct 470 ATNELVIRVQPDEAIYLKINNKVPGLGMRLDRSNLNLLYSARYSKE-IPDAYERLLLDAI 528
Query 69 KGDRQNFVRTDELREAWRIFTPLLKQIDEPGVKPEPYPYGSHGPESAY 116
+G+R+ F+R+DEL AW +FTPLLK+I+E PE YPYGS GP A+
Sbjct 529 EGERRLFIRSDELDAAWSLFTPLLKEIEEKKRIPEYYPYGSRGPVGAH 576
> At5g35790
Length=576
Score = 112 bits (280), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 60/117 (51%), Positives = 81/117 (69%), Gaps = 3/117 (2%)
Query 1 SFYPEEELSENELVIIVQPREAIYLKFYTKKPGLASGLQLTELDLSVMDRLQVDR-LPDA 59
SF + + NELVI VQP E IYL+ K PGL G++L DL+++ R + R +PDA
Sbjct 446 SFATNLDNATNELVIRVQPDEGIYLRINNKVPGL--GMRLDRSDLNLLYRSRYPREIPDA 503
Query 60 YERLLLDVIKGDRQNFVRTDELREAWRIFTPLLKQIDEPGVKPEPYPYGSHGPESAY 116
YERLLLD I+G+R+ F+R+DEL AW +FTP LK+++E + PE YPYGS GP A+
Sbjct 504 YERLLLDAIEGERRLFIRSDELDAAWDLFTPALKELEEKKIIPELYPYGSRGPVGAH 560
> At1g09420
Length=632
Score = 95.5 bits (236), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 48/110 (43%), Positives = 70/110 (63%), Gaps = 1/110 (0%)
Query 7 ELSENELVIIVQPREAIYLKFYTKKPGLASGLQLTELDLSVMDRLQVDRLPDAYERLLLD 66
+L NEL++ +P EAI +K K PGL L +EL+L DR + + +PD+YE L+ D
Sbjct 511 DLGTNELILRDEPDEAILVKINNKVPGLGLQLDASELNLLYKDRYKTE-VPDSYEHLIHD 569
Query 67 VIKGDRQNFVRTDELREAWRIFTPLLKQIDEPGVKPEPYPYGSHGPESAY 116
VI GD F+R+DE+ AW I +P+L++ID+ PE Y +G GP +AY
Sbjct 570 VIDGDNHLFMRSDEVAAAWNILSPVLEEIDKHHTAPELYEFGGRGPVAAY 619
> SPAC3A12.18
Length=447
Score = 77.4 bits (189), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 40/81 (49%), Positives = 49/81 (60%), Gaps = 0/81 (0%)
Query 10 ENELVIIVQPREAIYLKFYTKKPGLASGLQLTELDLSVMDRLQVDRLPDAYERLLLDVIK 69
NELVI VQP EAIY K K+PGL+ LT+LDL+ R + +L +AYE L LD
Sbjct 367 HNELVIRVQPDEAIYFKMNIKQPGLSEAPLLTDLDLTYSRRFKNMKLHEAYEALFLDAFA 426
Query 70 GDRQNFVRTDELREAWRIFTP 90
GD+ F R DEL AW + P
Sbjct 427 GDQSRFARIDELECAWSLVDP 447
> 7296516
Length=581
Score = 69.7 bits (169), Expect = 1e-12, Method: Composition-based stats.
Identities = 43/119 (36%), Positives = 66/119 (55%), Gaps = 8/119 (6%)
Query 1 SFYPEEELSENELVIIVQPREAIYLKFYTKKPGLASGLQLTELDLSVMDRLQVDRLPDAY 60
+F+ + NELV+ P E ++++ K+ G L+ +E++L V DR P
Sbjct 365 TFHSDSTDIRNELVLRSFPTEEVFMRMRLKRQGEDICLRESEINLRVDDRG-----PKGL 419
Query 61 ERL---LLDVIKGDRQNFVRTDELREAWRIFTPLLKQIDEPGVKPEPYPYGSHGPESAY 116
+ L LL+V +GD+ F+R+DE E WRIF+P+L ID +P Y +GS GP AY
Sbjct 420 QGLPGYLLNVFQGDQTLFMRSDEQCEIWRIFSPVLATIDSDRPRPLHYDFGSRGPLLAY 478
> SPCC794.01c
Length=475
Score = 68.2 bits (165), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 38/111 (34%), Positives = 60/111 (54%), Gaps = 3/111 (2%)
Query 9 SENELVIIVQPREAIYLKFYTKKPGLASGLQLTELDLSVMDRLQVDRLPDAYERLLLDVI 68
S N LV+ V P+E I LK + K+PG + + LD+ + + AYE ++ D I
Sbjct 358 SSNVLVLRVYPKEFIALKGHIKQPGFSRQIVPVTLDVKYPEAFPDTWIHKAYEVVIADAI 417
Query 69 KGDRQNFVRTDELREAWRIFTPLLKQIDEPGVKPEPYPYGS-HGPESAYTF 118
G +F+ DE+R +W+IF +L + + P PY +GS HGP++ F
Sbjct 418 NGKHTHFISDDEVRTSWKIFDDVLDTTGD--LSPLPYAFGSHHGPDATLEF 466
> SPAC3C7.13c
Length=473
Score = 57.0 bits (136), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 41/105 (39%), Positives = 54/105 (51%), Gaps = 9/105 (8%)
Query 13 LVIIVQPREAIYLKFYTKKPGLASGLQLTELDLSVMDRLQVDRL----PDAYERLLLDVI 68
L++ VQP E + L KP + + LQ + S+ Q L D YE L D I
Sbjct 361 LILHVQPEEFVNLTCTINKP-MTTDLQPIDAYASLNYNEQFKDLMKEKRDGYEILFEDAI 419
Query 69 KGDRQNFVRTDELREAWRIFTPLLKQIDEPGVKPEPYPYGSHGPE 113
+GD F+R DE+ AW+I+ +L D P KP PYP GS GPE
Sbjct 420 RGDPTKFIRYDEVEYAWKIWDEIL---DSPK-KPIPYPAGSDGPE 460
> Hs4758498_1
Length=557
Score = 36.2 bits (82), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 17/52 (32%), Positives = 27/52 (51%), Gaps = 2/52 (3%)
Query 58 DAYERLLLDVIKGDRQNFVRTDELREAWRIFTPLLKQIDEPGVKPEPYPYGS 109
DA+ LL + G + F+ T+ L +W +TPLL+ + P YP G+
Sbjct 456 DAHSVLLSHIFHGRKNFFITTENLLASWNFWTPLLESLAHKA--PRLYPGGA 505
> At1g64580
Length=1052
Score = 32.7 bits (73), Expect = 0.15, Method: Composition-based stats.
Identities = 15/35 (42%), Positives = 21/35 (60%), Gaps = 4/35 (11%)
Query 73 QNFVRTDELREAWRIFTPLLKQIDEPGVKPEPYPY 107
Q RTD+L+EAW +F L ++ GVKP+ Y
Sbjct 966 QGLCRTDKLKEAWCLFRSLTRK----GVKPDAIAY 996
> Hs14746694
Length=547
Score = 31.2 bits (69), Expect = 0.49, Method: Compositional matrix adjust.
Identities = 17/55 (30%), Positives = 27/55 (49%), Gaps = 0/55 (0%)
Query 53 VDRLPDAYERLLLDVIKGDRQNFVRTDELREAWRIFTPLLKQIDEPGVKPEPYPY 107
VD L +A L + ++F+ D + E W++ PLL + P PEPY +
Sbjct 168 VDDLLEALRSLCNRDTDMEVEDFIGVDSMYENWQVDRPLLCHLFVPFTPPEPYRF 222
> At1g05670_2
Length=687
Score = 30.0 bits (66), Expect = 0.98, Method: Composition-based stats.
Identities = 20/68 (29%), Positives = 33/68 (48%), Gaps = 14/68 (20%)
Query 52 QVDRLPDAYERLLLDVIKGDRQN----------FVRTDELREAWRIFTPLLKQIDEPGVK 101
Q+ R+ +A+ LLL +KG + + R EL + W+ L++ + G+K
Sbjct 204 QLGRIKEAHHLLLLMELKGYTPDVISYSTVVNGYCRFGELDKVWK----LIEVMKRKGLK 259
Query 102 PEPYPYGS 109
P Y YGS
Sbjct 260 PNSYIYGS 267
> YER012w
Length=198
Score = 29.6 bits (65), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 16/29 (55%), Positives = 17/29 (58%), Gaps = 1/29 (3%)
Query 90 PLLKQIDEPGVKPEPYPYGSHGPESAYTF 118
P L QID G K E PYG+HG YTF
Sbjct 114 PELYQIDYLGTKVE-LPYGAHGYSGFYTF 141
> Hs5453944
Length=753
Score = 28.5 bits (62), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 8/30 (26%), Positives = 16/30 (53%), Gaps = 0/30 (0%)
Query 64 LLDVIKGDRQNFVRTDELREAWRIFTPLLK 93
+ +I D F+ DE R+ W++F+ +
Sbjct 660 IFRIIDSDHSGFISLDEFRQTWKLFSSHMN 689
> Hs11641261
Length=960
Score = 28.1 bits (61), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 18/54 (33%), Positives = 26/54 (48%), Gaps = 2/54 (3%)
Query 24 YLKFYTKKPGLASGLQLTELDLSVMDRLQVDRLPDAYERLLLDVIK--GDRQNF 75
YL+ T P L GL E +MDR + + + +R LL K DRQ++
Sbjct 686 YLQHETSSPALLEGLSYLESFYHMMDRRNISDISENLKRYLLQYFKPVIDRQSW 739
> At5g62270
Length=404
Score = 26.9 bits (58), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 13/44 (29%), Positives = 25/44 (56%), Gaps = 0/44 (0%)
Query 58 DAYERLLLDVIKGDRQNFVRTDELREAWRIFTPLLKQIDEPGVK 101
+ Y+ LL ++ + +RQ D +++ + LLKQ+ EPG +
Sbjct 134 NEYDDLLKEIEQDNRQGRAFVDGIKQRMMEISVLLKQVKEPGAR 177
> Hs6005854
Length=299
Score = 26.9 bits (58), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 13/35 (37%), Positives = 18/35 (51%), Gaps = 0/35 (0%)
Query 32 PGLASGLQLTELDLSVMDRLQVDRLPDAYERLLLD 66
P + LQ+ + L V+ R LP Y+RL LD
Sbjct 93 PTGSKDLQMVNISLRVLSRPNAQELPSMYQRLGLD 127
Lambda K H
0.318 0.140 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1167969826
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40