bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0295_orf2
Length=171
Score E
Sequences producing significant alignments: (Bits) Value
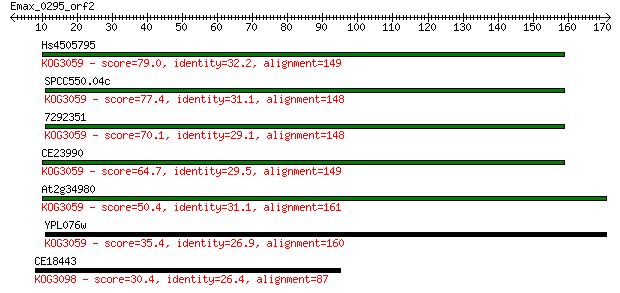
Hs4505795 79.0 4e-15
SPCC550.04c 77.4 1e-14
7292351 70.1 2e-12
CE23990 64.7 8e-11
At2g34980 50.4 2e-06
YPL076w 35.4 0.053
CE18443 30.4 1.8
> Hs4505795
Length=297
Score = 79.0 bits (193), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 48/156 (30%), Positives = 83/156 (53%), Gaps = 8/156 (5%)
Query 10 KWQKILWKKQPHPDCYVDETFLNTLMRNANLTRYMYSDLCKSTVVLTQHLSCILIFLLVY 69
KWQK+L+++QP PD YVD FL L +N + +Y Y + + V+ Q L + +F++++
Sbjct 13 KWQKVLYERQPFPDNYVDRRFLEELRKNIHARKYQYWAVVFESSVVIQQLCSVCVFVVIW 72
Query 70 RMIVKRSLAASTLVIFDVVSLPLGYALRWSLRPAPRAARQV-------LQSAIIVFGVLR 122
+ + LA L+ + S +GY L + L ++ L+SA++
Sbjct 73 WYMDEGLLAPHWLLGTGLASSLIGYVL-FDLIDGGEGRKKSGQTRWADLKSALVFITFTY 131
Query 123 ILAPVLQTLTQSFSDDTVISLTSICLLIHIPLNDYS 158
+PVL+TLT+S S DT+ +++ LL H+ DY
Sbjct 132 GFSPVLKTLTESVSTDTIYAMSVFMLLGHLIFFDYG 167
> SPCC550.04c
Length=324
Score = 77.4 bits (189), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 46/154 (29%), Positives = 85/154 (55%), Gaps = 6/154 (3%)
Query 11 WQKILWKKQPHPDCYVDETFLNTLMRNANLTRYMYSDLCKSTVVLTQHLSCILIFLLVYR 70
W+K+LW+KQ +PD ++DE+FLN L RN N+ + L ++ ++QHLS ++IF V+
Sbjct 38 WKKVLWRKQDYPDNFIDESFLNGLQRNVNIQVTDFWSLVADSLPVSQHLSSVVIFASVFV 97
Query 71 MIVKRSLAASTLVIFDVVSLPLGYALRWSLRPAPRAAR------QVLQSAIIVFGVLRIL 124
I + L+ + + VS + L + P R +++S I++ L L
Sbjct 98 SIYRNQLSCALVGFVSNVSAVAAFILWDFVLRKPCNNRTFPNYMGIVKSCILIVLTLAGL 157
Query 125 APVLQTLTQSFSDDTVISLTSICLLIHIPLNDYS 158
+P+L +LT+S S D+V ++ L ++ ++Y+
Sbjct 158 SPILMSLTKSTSPDSVWAIAVWLFLANVFFHEYT 191
> 7292351
Length=256
Score = 70.1 bits (170), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 43/154 (27%), Positives = 77/154 (50%), Gaps = 12/154 (7%)
Query 11 WQKILWKKQPHPDCYVDETFLNTLMRNANLTRYMYSDLCKSTVVLTQHLSCILIFLLVYR 70
W K L+ + +PD Y D +FL L N + Y + + VL +SCI FL++Y+
Sbjct 12 WVKNLYSNREYPDNYTDASFLKDLRTNLHCRIYTFGEAIAGITVLNNQISCITGFLILYQ 71
Query 71 MIVKRSLAASTLVIFDVVSLPLGY------ALRWSLRPAPRAARQVLQSAIIVFGVLRIL 124
+++ S++ +T+++ +GY +L W+L +++FG L
Sbjct 72 LMLSDSVSPTTILVPSCGITGIGYLCYRGRSLSWALLGEDSKTL----VTVVLFGYL--F 125
Query 125 APVLQTLTQSFSDDTVISLTSICLLIHIPLNDYS 158
+P+L TLTQ+ S DT+ ++T LL ++ Y
Sbjct 126 SPMLHTLTQAISTDTIYTMTFFVLLGNLIFGHYG 159
> CE23990
Length=282
Score = 64.7 bits (156), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 44/153 (28%), Positives = 75/153 (49%), Gaps = 6/153 (3%)
Query 10 KWQKILWKKQPHPDCYV--DETFLNTLMRNANLTRYMYSDLCKSTVVLTQHLSCILIFLL 67
WQKIL++KQP PD Y D FL L +N ++ Y Y + HL I ++ +
Sbjct 6 GWQKILYRKQPFPDNYSGGDAQFLKELRKNVSVVHYDYKSAVFGCMNFLTHLDMITMYFV 65
Query 68 VYRMIVKRSLAASTLVIFDVVSLPLGYALRWS--LRPAPRAARQVLQSAIIVFGVLRILA 125
++ I+ + + + +++ V SL + L + L P P A++ ++ +F
Sbjct 66 LFLNILHSNWSIN--ILYSVFSLTIVLYLFFCKFLIPNPANAKEHARTIFTLFIFAYAFT 123
Query 126 PVLQTLTQSFSDDTVISLTSICLLIHIPLNDYS 158
PV++TLT S S DT+ S + I + +DY
Sbjct 124 PVIRTLTTSISTDTIYSTSIITAIFSCFFHDYG 156
> At2g34980
Length=303
Score = 50.4 bits (119), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 50/168 (29%), Positives = 90/168 (53%), Gaps = 8/168 (4%)
Query 10 KWQKILWKKQ--PHPDCYVDETFLNTLMRNANLTRYMYSDLCKSTVVLTQHLSCILIFLL 67
KW+K+ + + D Y DE+FL ++ NAN+ R + K +V ++Q+L + + +L
Sbjct 14 KWRKVAYGGMQIGYDDNYTDESFLEEMVMNANVVRRDLLKVMKDSVSISQYLCIVALVVL 73
Query 68 VYRMIVKRSLAASTLVIFDVVSLPLGYAL----RWSLRPAPRAARQVLQSAIIVFGVLRI 123
V+ ++ SL ++L++ D+ L G+ + + R ++ + G L I
Sbjct 74 VWVHTLESSLDENSLLLLDLSLLASGFLILLLTEEKMLSLSLLLRYLVNISFFTTG-LYI 132
Query 124 LAPVLQTLTQSFSDDTVISLTSICLLIHIPLNDYS-YVYRNPETIDEP 170
LAP+ QTLT+S S D++ ++T LL+H+ L+DYS R P + P
Sbjct 133 LAPIYQTLTRSISSDSIWAVTVSLLLLHLFLHDYSGSTIRAPGALKTP 180
> YPL076w
Length=280
Score = 35.4 bits (80), Expect = 0.053, Method: Compositional matrix adjust.
Identities = 43/171 (25%), Positives = 76/171 (44%), Gaps = 22/171 (12%)
Query 11 WQKILWKKQPHPDCYVDETFLNTLMRNANLTRYMYSDLCKSTVVLTQHLSCILIFLLVYR 70
W+++LW KQ +PD Y D +F+ R + SD S Q L F+ Y+
Sbjct 6 WKRLLWLKQEYPDNYTDPSFIELRARQKAESN-QKSDRKLSEAARAQ---IRLDFISFYQ 61
Query 71 MIVKRSLAASTLVI-----FDVVSLPLGYAL------RWSLRPAPRAARQVLQSAIIVFG 119
I+ S T FD + + + R + P + V S II F
Sbjct 62 TILNTSFIYITFTYIYYYGFDPIPPTIFLSFITLIISRTKVDPLLSSFMDVKSSLIITFA 121
Query 120 VLRILAPVLQTLTQSFSDDTVISLTSICLLIHIPLNDYSYVYRNPETIDEP 170
+L L+PVL++L+++ + D++ +L+ L +I +V + ++ D+P
Sbjct 122 MLT-LSPVLKSLSKTTASDSIWTLSFWLTLWYI------FVISSTKSKDKP 165
> CE18443
Length=435
Score = 30.4 bits (67), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 23/87 (26%), Positives = 40/87 (45%), Gaps = 13/87 (14%)
Query 8 FGKWQKILWKKQPHPDCYVDETFLNTLMRNANLTRYMYSDLCKSTVVLTQHLSCILIFLL 67
+G ++KIL ++++ F L RN N+ Y Y DL ++ L +C+
Sbjct 16 YGSYKKIL---------FLEKNFAK-LQRNFNVVFYTYCDLYQAVCYLAYVTTCLFSPSF 65
Query 68 VYRMIVKRSLAASTLVIFDVVSLPLGY 94
+Y K +L S++ S PLG+
Sbjct 66 LYATSAKTTLLISSICF---TSFPLGF 89
Lambda K H
0.327 0.138 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2526689620
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40