bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
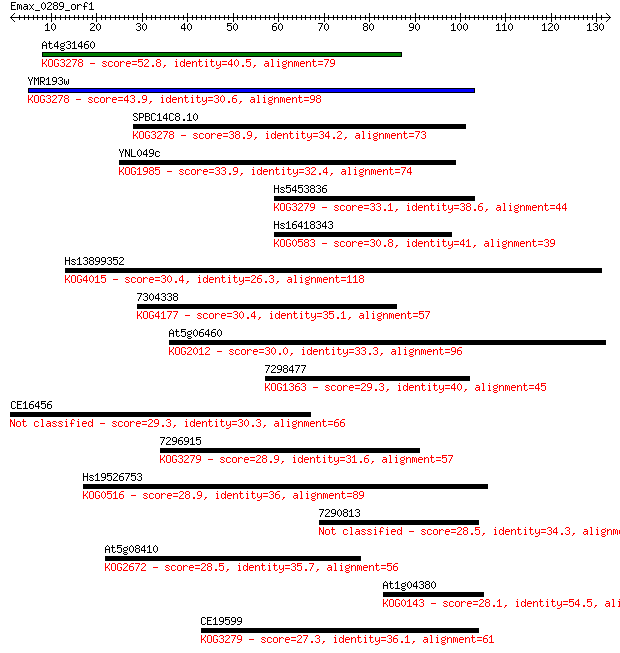
Query= Emax_0289_orf1
Length=132
Score E
Sequences producing significant alignments: (Bits) Value
At4g31460 52.8 2e-07
YMR193w 43.9 8e-05
SPBC14C8.10 38.9 0.002
YNL049c 33.9 0.094
Hs5453836 33.1 0.13
Hs16418343 30.8 0.76
Hs13899352 30.4 0.90
7304338 30.4 1.1
At5g06460 30.0 1.1
7298477 29.3 2.1
CE16456 29.3 2.3
7296915 28.9 2.5
Hs19526753 28.9 2.8
7290813 28.5 3.1
At5g08410 28.5 3.2
At1g04380 28.1 4.3
CE19599 27.3 7.7
> At4g31460
Length=212
Score = 52.8 bits (125), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 32/80 (40%), Positives = 48/80 (60%), Gaps = 2/80 (2%)
Query 8 GLFHDEDYNYYTKISF-SLRKHRRKLKPNVLKKDFESEGLNTVVKNLKVTTSAIHAIDDA 66
GL+ Y ++S K RR KPNV +K S ++ +K +KVTT A+ ID A
Sbjct 46 GLYAGRHIQYGNRVSEDGGNKSRRCWKPNVQEKRLFSYIFDSHIK-VKVTTHALRCIDKA 104
Query 67 GGLDEYLLRTPPEELRSVLG 86
GG+DEYLL+TP +++ + +G
Sbjct 105 GGIDEYLLKTPYQKMDTEMG 124
> YMR193w
Length=258
Score = 43.9 bits (102), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 30/98 (30%), Positives = 46/98 (46%), Gaps = 1/98 (1%)
Query 5 SQCGLFHDEDYNYYTKISFSLRKHRRKLKPNVLKKDFESEGLNTVVKNLKVTTSAIHAID 64
S GL+ + IS S K R+K PNV+KK SE LN + ++K+T + I
Sbjct 69 SNKGLYGGSFVQFGNNISESKAKTRKKWLPNVVKKGLWSETLNRKI-SIKMTAKVLKTIS 127
Query 65 DAGGLDEYLLRTPPEELRSVLGEKMKAVIRFYERNPEV 102
GG+D YL + ++ + K R +R E+
Sbjct 128 KEGGIDNYLTKEKSARIKELGPTGWKLRYRVLKRKDEI 165
> SPBC14C8.10
Length=176
Score = 38.9 bits (89), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 25/77 (32%), Positives = 43/77 (55%), Gaps = 14/77 (18%)
Query 28 HRRKLKPNVLKKDFESEGLNTVVKNLKVTTSAIHAIDDAGGLDEYLLRTPPEELRS--VL 85
H + L NVL++ F +L VT+ + ID GGLDEYL+++ P L+S +
Sbjct 60 HSKWLYSNVLEEKF----------HLYVTSRVLRTIDKEGGLDEYLVKSTPSRLKSLGLR 109
Query 86 GEKMKAVI--RFYERNP 100
G +++A++ + +NP
Sbjct 110 GVELRALVLHKLGAKNP 126
> YNL049c
Length=876
Score = 33.9 bits (76), Expect = 0.094, Method: Compositional matrix adjust.
Identities = 24/82 (29%), Positives = 42/82 (51%), Gaps = 16/82 (19%)
Query 25 LRKHRRKLKPNVLKKDF--------ESEGLNTVVKNLKVTTSAIHAIDDAGGLDEYLLRT 76
L KH L+P V+ D+ E+E L+ ++K++ T ++H + D GL ++
Sbjct 673 LSKHI-ALRPGVVPSDYRASALNRLETEPLHYLIKSIYPTVYSLHDMPDEVGLPDF---- 727
Query 77 PPEELRSVLGEKMKAVIRFYER 98
E ++VL E + A I +ER
Sbjct 728 ---EGKTVLPEPINATISLFER 746
> Hs5453836
Length=128
Score = 33.1 bits (74), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 17/46 (36%), Positives = 30/46 (65%), Gaps = 2/46 (4%)
Query 59 AIHAIDDAGGLDEYLLRTPPEELRSVLGEKMK--AVIRFYERNPEV 102
+ ID+A GLD Y+L+TP E+L S G ++K ++R ++P++
Sbjct 3 TLDLIDEAYGLDFYILKTPKEDLCSKFGMELKRGMLLRLARQDPQL 48
> Hs16418343
Length=268
Score = 30.8 bits (68), Expect = 0.76, Method: Compositional matrix adjust.
Identities = 16/39 (41%), Positives = 22/39 (56%), Gaps = 0/39 (0%)
Query 59 AIHAIDDAGGLDEYLLRTPPEELRSVLGEKMKAVIRFYE 97
AI ID GG +E++ R P EL+ V K +I+ YE
Sbjct 37 AIKVIDKMGGPEEFIQRFLPRELQIVRTLDHKNIIQVYE 75
> Hs13899352
Length=135
Score = 30.4 bits (67), Expect = 0.90, Method: Compositional matrix adjust.
Identities = 31/129 (24%), Positives = 51/129 (39%), Gaps = 22/129 (17%)
Query 13 EDYNYYTKISFSLRKHRRKLKPNVLKKDFESEGLNTVVKNLKVTTSAIHAIDDAGGLDEY 72
EDY IS ++RK LKP+ K+ E +G + V+ L + D +E
Sbjct 18 EDYLQALNISLAVRKIALLLKPD---KEIEHQGNHMTVRTLSTFRNYTVQFDVGVEFEE- 73
Query 73 LLRTPPEELRSVLGEKMKAVIRFYER-----------NPEVKEWALPWKSFASAYARGDP 121
+LRSV G K + ++ + E N + W + AR
Sbjct 74 -------DLRSVDGRKCQTIVTWEEEHLVCVQKGEVPNRGWRHWLEGEMLYLELTARDAV 126
Query 122 CQALYRHLR 130
C+ ++R +R
Sbjct 127 CEQVFRKVR 135
> 7304338
Length=1526
Score = 30.4 bits (67), Expect = 1.1, Method: Composition-based stats.
Identities = 20/64 (31%), Positives = 30/64 (46%), Gaps = 7/64 (10%)
Query 29 RRKLKPNVLKKDFESE-------GLNTVVKNLKVTTSAIHAIDDAGGLDEYLLRTPPEEL 81
R K PN L KD + G TV+++LK+ TS + G ++E L PE +
Sbjct 758 RHKANPNALTKDGNTALHIASNLGYVTVMESLKIVTSTSVINSNIGAIEEKLKVMTPELM 817
Query 82 RSVL 85
+ L
Sbjct 818 QETL 821
> At5g06460
Length=1077
Score = 30.0 bits (66), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 32/125 (25%), Positives = 54/125 (43%), Gaps = 29/125 (23%)
Query 36 VLKKDFESEGLNTVVKNLKVTTSAIHAIDDAGGLDEY---LLRT-----PPEELRSVLGE 87
V+ DFE + T+V + K TT + ++DDA +DE L+R P ++++ E
Sbjct 819 VIVPDFEPKKDATIVTDEKATTLSTASVDDAAVIDELNAKLVRCRMSLQPEFRMKAIQFE 878
Query 88 K----------MKAVIRFYERN---PEVKEW--------ALPWKSFASAYARGDPCQALY 126
K + + RN PEV + +P + ++A A G C +Y
Sbjct 879 KDDDTNYHMDMIAGLANMRARNYSVPEVDKLKAKFIAGRIIPAIATSTAMATGFVCLEMY 938
Query 127 RHLRG 131
+ L G
Sbjct 939 KVLDG 943
> 7298477
Length=464
Score = 29.3 bits (64), Expect = 2.1, Method: Composition-based stats.
Identities = 18/45 (40%), Positives = 24/45 (53%), Gaps = 6/45 (13%)
Query 57 TSAIHAIDDAGGLDEYLLRTPPEELRSVLGEKMKAVIRFYERNPE 101
TS + A + GG +E L T P LG+ MK + +YER PE
Sbjct 121 TSIVSAFVNLGGGNEARLVTDP------LGDVMKFIREYYERYPE 159
> CE16456
Length=689
Score = 29.3 bits (64), Expect = 2.3, Method: Composition-based stats.
Identities = 20/74 (27%), Positives = 31/74 (41%), Gaps = 15/74 (20%)
Query 1 PSAASQCGLFHDEDYNYYTKISFSLRKHRRKLKPNVLKKDF--------ESEGLNTVVKN 52
P ++ L HD Y +R RK+ PN+ KK F E ++T+ KN
Sbjct 544 PYNITENDLEHDRTY-------LEMRSQLRKMLPNIKKKIFILDSIPRVHPEQIDTIAKN 596
Query 53 LKVTTSAIHAIDDA 66
LK + I+ +
Sbjct 597 LKRKRKTMEEINKS 610
> 7296915
Length=302
Score = 28.9 bits (63), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 18/57 (31%), Positives = 29/57 (50%), Gaps = 1/57 (1%)
Query 34 PNVLKKDFESEGLNTVVKNLKVTTSAIHAIDDAGGLDEYLLRTPPEELRSVLGEKMK 90
PN+ + S L+ + ++ VT + I + G D YLL+ +LRS L K+K
Sbjct 112 PNLRRSVVHSHVLDCYM-SVVVTERTLEQIHECHGFDHYLLKNRACDLRSALALKLK 167
> Hs19526753
Length=3321
Score = 28.9 bits (63), Expect = 2.8, Method: Composition-based stats.
Identities = 32/104 (30%), Positives = 49/104 (47%), Gaps = 15/104 (14%)
Query 17 YYTKISFSLRKHRRKLKPNVLK-KDFESEGLN-----TVVKNLKVTTSAIHAIDDAGGLD 70
+ K+S SL H +L ++ K+ E+ + T + LK T SA + DD L+
Sbjct 2216 FKKKLSQSLPDHHEELHAEQMRCKELENAVGSWTDDLTQLSLLKDTLSAYISADDISILN 2275
Query 71 EY--LLRTPPEEL-------RSVLGEKMKAVIRFYERNPEVKEW 105
E LL+ EEL R +GE++ F E+N E+ EW
Sbjct 2276 ERVELLQRQWEELCHQLSLRRQQIGERLNEWAVFSEKNKELCEW 2319
> 7290813
Length=210
Score = 28.5 bits (62), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 12/35 (34%), Positives = 20/35 (57%), Gaps = 0/35 (0%)
Query 69 LDEYLLRTPPEELRSVLGEKMKAVIRFYERNPEVK 103
L + +R+P E R V+G + +R Y+RN + K
Sbjct 119 LRKQFMRSPDEISREVMGRDWEETVRTYKRNAQSK 153
> At5g08410
Length=477
Score = 28.5 bits (62), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 20/57 (35%), Positives = 28/57 (49%), Gaps = 1/57 (1%)
Query 22 SFSLRKHRRKLKPNVLKKDFESEGLNTVVKNLKVTTSAIHAID-DAGGLDEYLLRTP 77
S S+ KH + KP ++ K GL + LK + + AID D L +YL TP
Sbjct 203 SMSVLKHAKISKPGMITKTSIMLGLGETDEELKEAMADLRAIDVDILTLGQYLQPTP 259
> At1g04380
Length=345
Score = 28.1 bits (61), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 12/22 (54%), Positives = 16/22 (72%), Gaps = 0/22 (0%)
Query 83 SVLGEKMKAVIRFYERNPEVKE 104
SVL E V+RF+E +PEVK+
Sbjct 84 SVLEEIQNGVVRFHEEDPEVKK 105
> CE19599
Length=310
Score = 27.3 bits (59), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 22/68 (32%), Positives = 33/68 (48%), Gaps = 8/68 (11%)
Query 43 SEGLNTVVKNLKVTTSAIHAIDDAGGLDEYLLRTPPEELRSVLGEKMKAVI-------RF 95
SE L+ +K + VT A+ ID+ GLD Y+L + +L S +K + +
Sbjct 144 SEILDKYIK-VTVTERAMRLIDEHFGLDYYILESKEIDLDSKFANSLKREMLLTLTTESY 202
Query 96 YERNPEVK 103
YE N E K
Sbjct 203 YEENDEKK 210
Lambda K H
0.318 0.135 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1319765976
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40