bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0278_orf1
Length=165
Score E
Sequences producing significant alignments: (Bits) Value
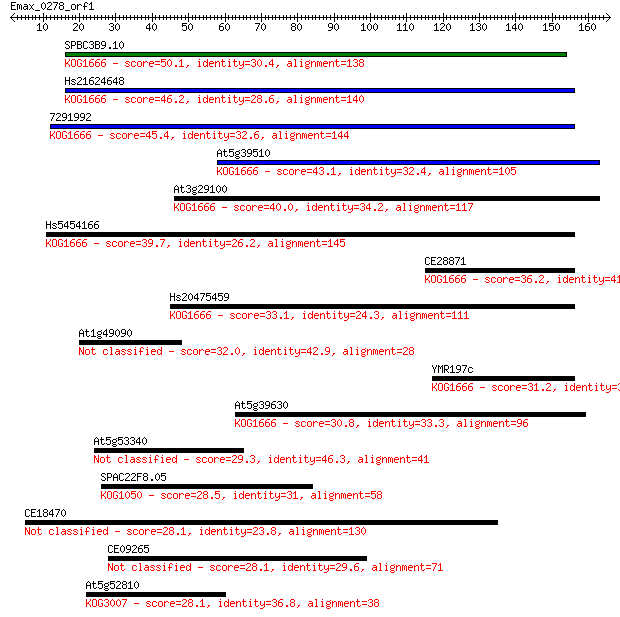
SPBC3B9.10 50.1 2e-06
Hs21624648 46.2 3e-05
7291992 45.4 5e-05
At5g39510 43.1 2e-04
At3g29100 40.0 0.002
Hs5454166 39.7 0.003
CE28871 36.2 0.028
Hs20475459 33.1 0.23
At1g49090 32.0 0.61
YMR197c 31.2 0.87
At5g39630 30.8 1.3
At5g53340 29.3 4.0
SPAC22F8.05 28.5 5.6
CE18470 28.1 7.2
CE09265 28.1 7.5
At5g52810 28.1 7.7
> SPBC3B9.10
Length=214
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 42/158 (26%), Positives = 84/158 (53%), Gaps = 21/158 (13%)
Query 16 YVAEFHSLRAELENMLDSAASRGDLFVATDSK--LREMKDCLGAFEMELSSMPPAQRS-- 71
Y E+ LRA++E L+ + G+ V + L E+ + +G E+E++ +P ++R
Sbjct 4 YEQEYRLLRADIEEKLNDLSKSGENSVIQSCQRLLNEIDEVIGQMEIEITGIPTSERGLV 63
Query 72 -----SHLNQLQTFR----DEFSRLQRRCLFATNR----GHEASASAELKRGTQALE--- 115
S+ + L+ +R +E + R+ LF NR G ++ + + T+ L+
Sbjct 64 NGRIRSYRSTLEEWRRHLKEEIGKSDRKALFG-NRDETSGDYIASDQDYDQRTRLLQGTN 122
Query 116 RLNSANIQLGNARKLAEETEEVGANILCNLYMQRESIQ 153
RL ++ +L ++++A ETE +GA+IL +L+ QR ++
Sbjct 123 RLEQSSQRLLESQRIANETEGIGASILRDLHGQRNQLE 160
> Hs21624648
Length=203
Score = 46.2 bits (108), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 40/159 (25%), Positives = 73/159 (45%), Gaps = 22/159 (13%)
Query 16 YVAEFHSLRAELENMLDSAA-----SRGDLFVATDSKLREMKDCLGAFEMELSSMPPAQR 70
Y +F L AE+ + + + + + +L E K+ L ++E+ +PP R
Sbjct 8 YEQDFAVLTAEITSKIARVPRLPPDEKKQMVANVEKQLEEAKELLEQMDLEVREIPPQSR 67
Query 71 SSHLNQLQTFRDEFSRLQ--------------RRCLFATNRGHEASASAELKRGTQALER 116
+ N++++++ E +L+ R L + + A L T+ LER
Sbjct 68 GMYSNRMRSYKQEMGKLETDFKRSRIAYSDEVRNELLGDDGNSSENQRAHLLDNTERLER 127
Query 117 LNSANIQLGNARKLAEETEEVGANILCNLYMQRESIQRA 155
+S ++ G ++A ETE++G +L NL RE IQRA
Sbjct 128 -SSRRLEAGY--QIAVETEQIGQEMLENLSHDREKIQRA 163
> 7291992
Length=230
Score = 45.4 bits (106), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 47/167 (28%), Positives = 79/167 (47%), Gaps = 23/167 (13%)
Query 12 MLNEYVAEFHSLRAELENML------DSAASRGDLFVATDSKLREMKDCLGAFEMELSSM 65
+L +Y ++ +L AE+ + ++ + R DL DS L E ++ L +E+ +
Sbjct 3 LLEQYEQQYATLIAEITAHIGRLQQQNNNSERHDLCSKIDSSLPEAQELLEQMGLEVREL 62
Query 66 PPAQRSSHLNQLQTFRDEFSRLQRRCLFATNRGH-EASASAELKRG---------TQALE 115
P RSS +LQ + E RLQ ++ +A+ L G T +
Sbjct 63 NPGLRSSFNGKLQVAQAELKRLQAEYRLTKDKQRSQANTFTTLDLGDSYEDVSISTDQRQ 122
Query 116 RL--NSANIQ-----LGNARKLAEETEEVGANILCNLYMQRESIQRA 155
RL NS I+ L ++A ETE++GA +L +L+ QRE++Q A
Sbjct 123 RLLDNSERIERTGNRLTEGYRVALETEQLGAQVLNDLHHQRETLQGA 169
> At5g39510
Length=221
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 34/123 (27%), Positives = 61/123 (49%), Gaps = 18/123 (14%)
Query 58 FEMELSSMPPAQRSSHLNQLQTFRDEFSRLQ---------------RRCLFATNRGHEAS 102
++E ++PP +SS L +L+ F+ + + + R L +
Sbjct 55 MDLEARTLPPNLKSSLLVKLREFKSDLNNFKTEVKRITSGQLNAAARDELLEAGMADTKT 114
Query 103 ASAELK-RGTQALERLNSANIQLGNARKLAEETEEVGANILCNLYMQRESIQRAN--LHA 159
ASA+ + R + ERL ++ ++R+ ETEE+G +IL +L+ QR+S+ RA+ LH
Sbjct 115 ASADQRARLMMSTERLGRTTDRVKDSRRTMMETEEIGVSILQDLHGQRQSLLRAHETLHG 174
Query 160 EAD 162
D
Sbjct 175 VDD 177
> At3g29100
Length=221
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 40/135 (29%), Positives = 63/135 (46%), Gaps = 18/135 (13%)
Query 46 SKLREMKDCLGAFEMELSSMPPAQRSSHL-------NQLQTFRDEFSRLQRRCLFATNRG 98
S + E + + ++E ++PP +SS L + L F+ E R+ L AT R
Sbjct 43 SGVEEAEALVKKMDLEARNLPPNVKSSLLVKLREYKSDLNNFKTEVKRITSGNLNATARD 102
Query 99 H--EASASAELKRGTQALERLNSANIQLG-------NARKLAEETEEVGANILCNLYMQR 149
EA + L RL + LG ++R+ ETEE+G +IL +L+ QR
Sbjct 103 ELLEAGMADTLTASADQRSRLMMSTDHLGRTTDRIKDSRRTILETEELGVSILQDLHGQR 162
Query 150 ESIQRAN--LHAEAD 162
+S+ RA+ LH D
Sbjct 163 QSLLRAHETLHGVDD 177
> Hs5454166
Length=232
Score = 39.7 bits (91), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 38/166 (22%), Positives = 72/166 (43%), Gaps = 21/166 (12%)
Query 11 EMLNEYVAEFH-SLRAELENMLDSAAS--RGDLFVATDSKLREMKDCLGAFEMELSSMPP 67
E L+E H L+ E +L +A + + L D K +E + L E EL P
Sbjct 12 EKLHEIFRGLHEDLQGVPERLLGTAGTEEKKKLIRDFDEKQQEANETLAEMEEELRYAPL 71
Query 68 AQRSSHLNQLQTFRDEFSRLQRRC------------------LFATNRGHEASASAELKR 109
+ R+ +++L+ +R + ++L R ++A H ++
Sbjct 72 SFRNPMMSKLRNYRKDLAKLHREVRSTPLTATPGGRGDMKYGIYAVENEHMNRLQSQRAM 131
Query 110 GTQALERLNSANIQLGNARKLAEETEEVGANILCNLYMQRESIQRA 155
Q E LN A + + ++A ET+++G+ I+ L QR+ ++R
Sbjct 132 LLQGTESLNRATQSIERSHRIATETDQIGSEIIEELGEQRDQLERT 177
> CE28871
Length=224
Score = 36.2 bits (82), Expect = 0.028, Method: Compositional matrix adjust.
Identities = 17/41 (41%), Positives = 29/41 (70%), Gaps = 0/41 (0%)
Query 115 ERLNSANIQLGNARKLAEETEEVGANILCNLYMQRESIQRA 155
+RL + ++ +A ++A ETE++GA +L NL QRE+I R+
Sbjct 133 QRLERSTRKVQDAHRIAVETEQIGAEMLSNLASQRETIGRS 173
> Hs20475459
Length=231
Score = 33.1 bits (74), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 27/129 (20%), Positives = 55/129 (42%), Gaps = 18/129 (13%)
Query 45 DSKLREMKDCLGAFEMELSSMPPAQRSSHLNQLQTFRDEFSRLQRRC------------- 91
D K +E + L E EL P + ++ ++L+ +R + ++L R
Sbjct 48 DEKQQEANEMLAGMEEELRYAPLSFHNTMTSKLRNYRKDLAKLHREVRSTPLTATPGGRG 107
Query 92 -----LFATNRGHEASASAELKRGTQALERLNSANIQLGNARKLAEETEEVGANILCNLY 146
++A H ++ Q E LN A + + ++A ET+++G+ + L
Sbjct 108 DMKYDIYAVENEHMNRLQSQRAMLLQGPENLNRATQSIERSHQIATETDQIGSETIEELG 167
Query 147 MQRESIQRA 155
QR+ ++R
Sbjct 168 EQRDHLERT 176
> At1g49090
Length=372
Score = 32.0 bits (71), Expect = 0.61, Method: Compositional matrix adjust.
Identities = 12/28 (42%), Positives = 18/28 (64%), Gaps = 0/28 (0%)
Query 20 FHSLRAELENMLDSAASRGDLFVATDSK 47
+H +R ELE+ L S GD+F+ T +K
Sbjct 169 YHQIRDELEDQLGKTVSIGDVFIKTHTK 196
> YMR197c
Length=217
Score = 31.2 bits (69), Expect = 0.87, Method: Compositional matrix adjust.
Identities = 14/39 (35%), Positives = 26/39 (66%), Gaps = 0/39 (0%)
Query 117 LNSANIQLGNARKLAEETEEVGANILCNLYMQRESIQRA 155
L + +L +A ++A ETE +G+ I+ +L QRE+++ A
Sbjct 127 LQKSGDRLKDASRIANETEGIGSQIMMDLRSQRETLENA 165
> At5g39630
Length=207
Score = 30.8 bits (68), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 32/113 (28%), Positives = 52/113 (46%), Gaps = 20/113 (17%)
Query 63 SSMPPAQRSSHLNQLQTFRDEFSRLQ---------------RRCLFATNRGHEASASAEL 107
S++PP +S L +L+ + RL+ R + + A + L
Sbjct 60 SNLPPNIKSILLEKLKESKSSLKRLRNEIKRNTSENLKVTTREEVLEAEKADLADQRSRL 119
Query 108 KRGTQALERLNSANIQLGNARKLAEETEEVGANILCNLYMQRESIQ--RANLH 158
+ T+ L R + + + RKL E TE +G +IL NL Q+ES+Q +A LH
Sbjct 120 MKSTEGLVR--TREMIKDSQRKLLE-TENIGISILENLQRQKESLQNSQAMLH 169
> At5g53340
Length=362
Score = 29.3 bits (64), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 19/46 (41%), Positives = 22/46 (47%), Gaps = 5/46 (10%)
Query 24 RAELENMLDSAASRGDLFVATDSKL-----REMKDCLGAFEMELSS 64
R L N LD +G ++ D L RE K L A EMELSS
Sbjct 42 RVHLINELDRVTGQGKSAISVDDTLKIIACREQKKTLAALEMELSS 87
> SPAC22F8.05
Length=891
Score = 28.5 bits (62), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 18/60 (30%), Positives = 33/60 (55%), Gaps = 2/60 (3%)
Query 26 ELENMLDSAASRGDLFVATDS--KLREMKDCLGAFEMELSSMPPAQRSSHLNQLQTFRDE 83
E++ +L+ + ++FV D +R +++ L AFE L P Q+++ L Q TF +E
Sbjct 405 EVKEVLEKRYANLNIFVGCDKMDPIRGIREKLLAFEQFLYDNPEYQKNTILIQTSTFTEE 464
> CE18470
Length=1475
Score = 28.1 bits (61), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 31/133 (23%), Positives = 62/133 (46%), Gaps = 11/133 (8%)
Query 5 GISILVEMLNEYVAE-FHSLRAELENMLDSAASRGDLFVATDSKLREMKDCLGAFEMELS 63
+ +++E L E + +LRAEL+ + DL + + K++E++ + E S
Sbjct 965 AVQVVMEALKSEQGESYEALRAELDAAVQEKGRSSDLVTSLEGKIQELETAI-----ESS 1019
Query 64 SMPPAQRSSHLNQLQTFRDEFSRLQRRC--LFATNRGHEASASAELKRGTQALERLNSAN 121
+ Q+S +Q F D+ S L+ + L + N E + + + ++ +L SAN
Sbjct 1020 TAENVQKSKT---IQDFTDKVSLLESQICELKSQNEQMEIDTNLNMDQLSEMSSQLESAN 1076
Query 122 IQLGNARKLAEET 134
+L + + ET
Sbjct 1077 AELIELTRTSAET 1089
> CE09265
Length=599
Score = 28.1 bits (61), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 21/71 (29%), Positives = 30/71 (42%), Gaps = 8/71 (11%)
Query 28 ENMLDSAASRGDLFVATDSKLREMKDCLGAFEMELSSMPPAQRSSHLNQLQTFRDEFSRL 87
EN+ D+ RG++ D K FE+ L SM P +S +TF L
Sbjct 512 ENIADALTIRGNIIRQLDDK--------SPFEVTLDSMIPVAATSRREAARTFYTVLELL 563
Query 88 QRRCLFATNRG 98
+ R + AT R
Sbjct 564 KERKIKATQRA 574
> At5g52810
Length=325
Score = 28.1 bits (61), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 14/38 (36%), Positives = 21/38 (55%), Gaps = 0/38 (0%)
Query 22 SLRAELENMLDSAASRGDLFVATDSKLREMKDCLGAFE 59
S E++ D+A RG +FV D+ + E + GAFE
Sbjct 231 SFSHEMKECDDNAIQRGSVFVDNDTAMIEAGELAGAFE 268
Lambda K H
0.316 0.128 0.344
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2353551590
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40