bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0224_orf1
Length=132
Score E
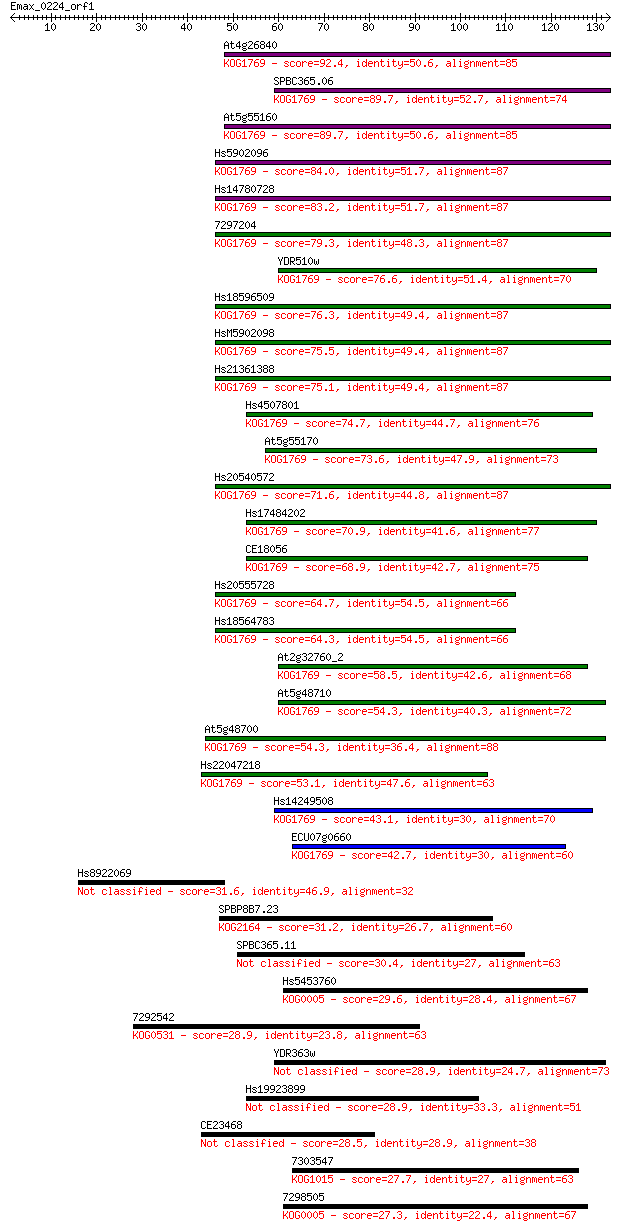
Sequences producing significant alignments: (Bits) Value
At4g26840 92.4 2e-19
SPBC365.06 89.7 1e-18
At5g55160 89.7 1e-18
Hs5902096 84.0 7e-17
Hs14780728 83.2 1e-16
7297204 79.3 2e-15
YDR510w 76.6 1e-14
Hs18596509 76.3 1e-14
HsM5902098 75.5 2e-14
Hs21361388 75.1 3e-14
Hs4507801 74.7 4e-14
At5g55170 73.6 1e-13
Hs20540572 71.6 3e-13
Hs17484202 70.9 6e-13
CE18056 68.9 3e-12
Hs20555728 64.7 4e-11
Hs18564783 64.3 5e-11
At2g32760_2 58.5 3e-09
At5g48710 54.3 5e-08
At5g48700 54.3 6e-08
Hs22047218 53.1 1e-07
Hs14249508 43.1 1e-04
ECU07g0660 42.7 2e-04
Hs8922069 31.6 0.38
SPBP8B7.23 31.2 0.50
SPBC365.11 30.4 0.99
Hs5453760 29.6 1.4
7292542 28.9 2.5
YDR363w 28.9 2.5
Hs19923899 28.9 2.6
CE23468 28.5 3.8
7303547 27.7 6.5
7298505 27.3 8.1
> At4g26840
Length=100
Score = 92.4 bits (228), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 43/85 (50%), Positives = 63/85 (74%), Gaps = 2/85 (2%)
Query 48 EEKKDGENAEKEHMQLKVRSPDGSEVYFKIKRRTKLEKLMTAYCNRLGQSIDSVRFLYDG 107
E+KK G+ H+ LKV+ DG+EV+F+IKR T+L+KLM AYC+R ++S+ FL+DG
Sbjct 7 EDKKPGDGGA--HINLKVKGQDGNEVFFRIKRSTQLKKLMNAYCDRQSVDMNSIAFLFDG 64
Query 108 ERVKPEKTPMDLGIEDGDVIDAMVQ 132
R++ E+TP +L +EDGD IDAM+
Sbjct 65 RRLRAEQTPDELDMEDGDEIDAMLH 89
> SPBC365.06
Length=117
Score = 89.7 bits (221), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 39/74 (52%), Positives = 58/74 (78%), Gaps = 0/74 (0%)
Query 59 EHMQLKVRSPDGSEVYFKIKRRTKLEKLMTAYCNRLGQSIDSVRFLYDGERVKPEKTPMD 118
EH+ LKV D +EV+FKIK+ T+ KLM YC R G+S++S+RFL DGER++P++TP +
Sbjct 34 EHINLKVVGQDNNEVFFKIKKTTEFSKLMKIYCARQGKSMNSLRFLVDGERIRPDQTPAE 93
Query 119 LGIEDGDVIDAMVQ 132
L +EDGD I+A+++
Sbjct 94 LDMEDGDQIEAVLE 107
> At5g55160
Length=103
Score = 89.7 bits (221), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 43/85 (50%), Positives = 62/85 (72%), Gaps = 3/85 (3%)
Query 48 EEKKDGENAEKEHMQLKVRSPDGSEVYFKIKRRTKLEKLMTAYCNRLGQSIDSVRFLYDG 107
E+KK + A H+ LKV+ DG+EV+F+IKR T+L+KLM AYC+R +S+ FL+DG
Sbjct 7 EDKKPDQGA---HINLKVKGQDGNEVFFRIKRSTQLKKLMNAYCDRQSVDFNSIAFLFDG 63
Query 108 ERVKPEKTPMDLGIEDGDVIDAMVQ 132
R++ E+TP +L +EDGD IDAM+
Sbjct 64 RRLRAEQTPDELEMEDGDEIDAMLH 88
> Hs5902096
Length=103
Score = 84.0 bits (206), Expect = 7e-17, Method: Compositional matrix adjust.
Identities = 45/88 (51%), Positives = 54/88 (61%), Gaps = 1/88 (1%)
Query 46 MSEEK-KDGENAEKEHMQLKVRSPDGSEVYFKIKRRTKLEKLMTAYCNRLGQSIDSVRFL 104
MSEEK K+G E +H+ LKV DGS V FKIKR T L KLM AYC R G S+ +RF
Sbjct 1 MSEEKPKEGVKTENDHINLKVAGQDGSVVQFKIKRHTSLSKLMKAYCERQGLSMRQIRFR 60
Query 105 YDGERVKPEKTPMDLGIEDGDVIDAMVQ 132
+DG+ + TP L +ED D ID Q
Sbjct 61 FDGQPINETDTPAQLRMEDEDTIDVFQQ 88
> Hs14780728
Length=103
Score = 83.2 bits (204), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 45/88 (51%), Positives = 54/88 (61%), Gaps = 1/88 (1%)
Query 46 MSEEK-KDGENAEKEHMQLKVRSPDGSEVYFKIKRRTKLEKLMTAYCNRLGQSIDSVRFL 104
MSEEK K+G E +H+ LKV DGS V FKIKR T L KLM AYC R G S+ +RF
Sbjct 1 MSEEKPKEGVKTENDHINLKVAGQDGSVVQFKIKRHTPLSKLMKAYCERQGLSMRQIRFR 60
Query 105 YDGERVKPEKTPMDLGIEDGDVIDAMVQ 132
+DG+ + TP L +ED D ID Q
Sbjct 61 FDGQPINETDTPAQLEMEDEDTIDVFQQ 88
> 7297204
Length=90
Score = 79.3 bits (194), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 42/87 (48%), Positives = 54/87 (62%), Gaps = 3/87 (3%)
Query 46 MSEEKKDGENAEKEHMQLKVRSPDGSEVYFKIKRRTKLEKLMTAYCNRLGQSIDSVRFLY 105
MS+EKK GE EH+ LKV D + V FKIK+ T L KLM AYC+R G S+ VRF +
Sbjct 1 MSDEKKGGET---EHINLKVLGQDNAVVQFKIKKHTPLRKLMNAYCDRAGLSMQVVRFRF 57
Query 106 DGERVKPEKTPMDLGIEDGDVIDAMVQ 132
DG+ + TP L +E+GD I+ Q
Sbjct 58 DGQPINENDTPTSLEMEEGDTIEVYQQ 84
> YDR510w
Length=101
Score = 76.6 bits (187), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 36/70 (51%), Positives = 52/70 (74%), Gaps = 1/70 (1%)
Query 60 HMQLKVRSPDGSEVYFKIKRRTKLEKLMTAYCNRLGQSIDSVRFLYDGERVKPEKTPMDL 119
H+ LKV S SE++FKIK+ T L +LM A+ R G+ +DS+RFLYDG R++ ++TP DL
Sbjct 23 HINLKV-SDGSSEIFFKIKKTTPLRRLMEAFAKRQGKEMDSLRFLYDGIRIQADQTPEDL 81
Query 120 GIEDGDVIDA 129
+ED D+I+A
Sbjct 82 DMEDNDIIEA 91
> Hs18596509
Length=95
Score = 76.3 bits (186), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 43/89 (48%), Positives = 54/89 (60%), Gaps = 2/89 (2%)
Query 46 MSEEK-KDGENAEK-EHMQLKVRSPDGSEVYFKIKRRTKLEKLMTAYCNRLGQSIDSVRF 103
M++EK K+G E +H+ LKV DGS V FKIKR T L KLM AYC R G S+ +RF
Sbjct 1 MADEKPKEGVKTENNDHINLKVAGQDGSVVQFKIKRHTPLSKLMKAYCERQGLSVRQIRF 60
Query 104 LYDGERVKPEKTPMDLGIEDGDVIDAMVQ 132
+DG+ + TP L +ED D ID Q
Sbjct 61 RFDGQPINETDTPAQLEMEDEDTIDVFQQ 89
> HsM5902098
Length=95
Score = 75.5 bits (184), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 43/89 (48%), Positives = 54/89 (60%), Gaps = 2/89 (2%)
Query 46 MSEEK-KDGENAEK-EHMQLKVRSPDGSEVYFKIKRRTKLEKLMTAYCNRLGQSIDSVRF 103
M++EK K+G E +H+ LKV DGS V FKIKR T L KLM AYC R G S+ +RF
Sbjct 1 MADEKPKEGVKTENNDHINLKVAGQDGSVVQFKIKRHTPLSKLMKAYCERQGLSMRQIRF 60
Query 104 LYDGERVKPEKTPMDLGIEDGDVIDAMVQ 132
+DG+ + TP L +ED D ID Q
Sbjct 61 RFDGQPINETDTPAQLEMEDEDTIDVFQQ 89
> Hs21361388
Length=95
Score = 75.1 bits (183), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 43/89 (48%), Positives = 53/89 (59%), Gaps = 2/89 (2%)
Query 46 MSEEK-KDGENAEKE-HMQLKVRSPDGSEVYFKIKRRTKLEKLMTAYCNRLGQSIDSVRF 103
M++EK K+G E H+ LKV DGS V FKIKR T L KLM AYC R G S+ +RF
Sbjct 1 MADEKPKEGVKTENNNHINLKVAGQDGSVVQFKIKRHTPLSKLMKAYCERQGLSMRQIRF 60
Query 104 LYDGERVKPEKTPMDLGIEDGDVIDAMVQ 132
+DG+ + TP L +ED D ID Q
Sbjct 61 RFDGQPINETDTPAQLEMEDEDTIDVFQQ 89
> Hs4507801
Length=101
Score = 74.7 bits (182), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 34/76 (44%), Positives = 54/76 (71%), Gaps = 0/76 (0%)
Query 53 GENAEKEHMQLKVRSPDGSEVYFKIKRRTKLEKLMTAYCNRLGQSIDSVRFLYDGERVKP 112
G+ E E+++LKV D SE++FK+K T L+KL +YC R G ++S+RFL++G+R+
Sbjct 14 GDKKEGEYIKLKVIGQDSSEIHFKVKMTTHLKKLKESYCQRQGVPMNSLRFLFEGQRIAD 73
Query 113 EKTPMDLGIEDGDVID 128
TP +LG+E+ DVI+
Sbjct 74 NHTPKELGMEEEDVIE 89
> At5g55170
Length=111
Score = 73.6 bits (179), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 35/73 (47%), Positives = 51/73 (69%), Gaps = 0/73 (0%)
Query 57 EKEHMQLKVRSPDGSEVYFKIKRRTKLEKLMTAYCNRLGQSIDSVRFLYDGERVKPEKTP 116
++ H+ LKV+S DG EV FK K+ L+KLM YC+R G +D+ F+++G R+ +TP
Sbjct 14 QEAHVILKVKSQDGDEVLFKNKKSAPLKKLMYVYCDRRGLKLDAFAFIFNGARIGGLETP 73
Query 117 MDLGIEDGDVIDA 129
+L +EDGDVIDA
Sbjct 74 DELDMEDGDVIDA 86
> Hs20540572
Length=95
Score = 71.6 bits (174), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 39/89 (43%), Positives = 53/89 (59%), Gaps = 2/89 (2%)
Query 46 MSEEKKDG--ENAEKEHMQLKVRSPDGSEVYFKIKRRTKLEKLMTAYCNRLGQSIDSVRF 103
M++EK + ++ +H LKV DGS V FKIKR T L KLM AYC + G S+ +RF
Sbjct 1 MADEKPNQGVKSENNDHTNLKVAGQDGSVVQFKIKRHTPLSKLMKAYCKQQGLSMRQIRF 60
Query 104 LYDGERVKPEKTPMDLGIEDGDVIDAMVQ 132
+DG+ +K TP L +E+ D ID Q
Sbjct 61 RFDGQPIKETDTPAQLEMENEDTIDVFQQ 89
> Hs17484202
Length=101
Score = 70.9 bits (172), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 32/77 (41%), Positives = 53/77 (68%), Gaps = 0/77 (0%)
Query 53 GENAEKEHMQLKVRSPDGSEVYFKIKRRTKLEKLMTAYCNRLGQSIDSVRFLYDGERVKP 112
G+ + E ++L+V D SE++FK+K T L+KL +YC R G ++S+RFL++G+R+
Sbjct 14 GDKIKDEDIKLRVIGQDSSEIHFKVKMTTPLKKLKKSYCQRQGVPVNSLRFLFEGQRIAD 73
Query 113 EKTPMDLGIEDGDVIDA 129
TP +LG+E+ DVI+
Sbjct 74 NHTPEELGMEEEDVIEV 90
> CE18056
Length=91
Score = 68.9 bits (167), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 32/75 (42%), Positives = 52/75 (69%), Gaps = 2/75 (2%)
Query 53 GENAEKEHMQLKVRSPDGSEVYFKIKRRTKLEKLMTAYCNRLGQSIDSVRFLYDGERVKP 112
G+NAE ++++KV D +EV+F++K T + KL +Y +R G +++S+RFL+DG R+
Sbjct 9 GDNAE--YIKIKVVGQDSNEVHFRVKYGTSMAKLKKSYADRTGVAVNSLRFLFDGRRIND 66
Query 113 EKTPMDLGIEDGDVI 127
+ TP L +ED DVI
Sbjct 67 DDTPKTLEMEDDDVI 81
> Hs20555728
Length=129
Score = 64.7 bits (156), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 36/68 (52%), Positives = 45/68 (66%), Gaps = 2/68 (2%)
Query 46 MSEEK-KDGENAEK-EHMQLKVRSPDGSEVYFKIKRRTKLEKLMTAYCNRLGQSIDSVRF 103
M+EEK K+G E +H+ LKV DGS V FKIKR T L KLM AYC R G S+ +RF
Sbjct 1 MAEEKPKEGVKTENNDHINLKVAGQDGSVVQFKIKRHTPLSKLMKAYCERQGLSMRQIRF 60
Query 104 LYDGERVK 111
+DG+ +K
Sbjct 61 RFDGQPMK 68
> Hs18564783
Length=126
Score = 64.3 bits (155), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 36/68 (52%), Positives = 45/68 (66%), Gaps = 2/68 (2%)
Query 46 MSEEK-KDGENAEK-EHMQLKVRSPDGSEVYFKIKRRTKLEKLMTAYCNRLGQSIDSVRF 103
M+EEK K+G E +H+ LKV DGS V FKIKR T L KLM AYC R G S+ +RF
Sbjct 1 MAEEKPKEGVKTENNDHINLKVAGQDGSVVQFKIKRHTPLSKLMKAYCERQGLSMRQIRF 60
Query 104 LYDGERVK 111
+DG+ +K
Sbjct 61 RFDGQPMK 68
> At2g32760_2
Length=94
Score = 58.5 bits (140), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 29/70 (41%), Positives = 43/70 (61%), Gaps = 2/70 (2%)
Query 60 HMQ--LKVRSPDGSEVYFKIKRRTKLEKLMTAYCNRLGQSIDSVRFLYDGERVKPEKTPM 117
HM+ LKV++ G+E +KI L+KLM+AYC + SVRF+Y+G +K +TP
Sbjct 11 HMKVTLKVKNQQGAEDLYKIGTHAHLKKLMSAYCTKRNLDYSSVRFVYNGREIKARQTPA 70
Query 118 DLGIEDGDVI 127
L +E+ D I
Sbjct 71 QLHMEEEDEI 80
> At5g48710
Length=114
Score = 54.3 bits (129), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 29/73 (39%), Positives = 43/73 (58%), Gaps = 1/73 (1%)
Query 60 HMQLKVRSPDGSEV-YFKIKRRTKLEKLMTAYCNRLGQSIDSVRFLYDGERVKPEKTPMD 118
H+ LKV+ D + F ++R KL K+M Y G ++ RFL+DG R++ TP +
Sbjct 27 HVTLKVKGQDEEDFRVFWVRRNAKLLKMMELYTKMRGIEWNTFRFLFDGSRIREYHTPDE 86
Query 119 LGIEDGDVIDAMV 131
L +DGD IDAM+
Sbjct 87 LERKDGDEIDAML 99
> At5g48700
Length=117
Score = 54.3 bits (129), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 32/90 (35%), Positives = 51/90 (56%), Gaps = 2/90 (2%)
Query 44 LKMSEEKKDGENAEKEHMQLKVRSPDGSEV-YFKIKRRTKLEKLMTAYCNRLGQSIDSVR 102
+KM EK+ +E H+ L V+ D V F+++R+ +L KLM Y G ++ R
Sbjct 14 VKMEGEKRKDVESESTHVTLNVKGQDEEGVKVFRVRRKARLLKLMEYYAKMRGIEWNTFR 73
Query 103 FLYD-GERVKPEKTPMDLGIEDGDVIDAMV 131
FL D G R++ T D+ ++DGD IDA++
Sbjct 74 FLSDDGSRIREYHTADDMELKDGDQIDALL 103
> Hs22047218
Length=265
Score = 53.1 bits (126), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 30/65 (46%), Positives = 39/65 (60%), Gaps = 2/65 (3%)
Query 43 FLKMSEEK-KDGENAEK-EHMQLKVRSPDGSEVYFKIKRRTKLEKLMTAYCNRLGQSIDS 100
L +++EK K+G AE +H L V DGS + FKIKR T L KLM AYC R G S+
Sbjct 100 MLTIADEKPKEGVKAENHDHTNLTVAGKDGSVMQFKIKRHTPLSKLMKAYCERQGLSMRQ 159
Query 101 VRFLY 105
+R +
Sbjct 160 IRLQF 164
> Hs14249508
Length=138
Score = 43.1 bits (100), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 21/71 (29%), Positives = 40/71 (56%), Gaps = 1/71 (1%)
Query 59 EHMQLKVRSPDGSEVY-FKIKRRTKLEKLMTAYCNRLGQSIDSVRFLYDGERVKPEKTPM 117
+ +QL+V+ + + + R + L+ LM+ Y +G S + F +DG ++ + P
Sbjct 65 QQLQLRVQGKEKHQTLEVSLSRDSPLKTLMSHYEEAMGLSGRKLSFFFDGTKLSGRELPA 124
Query 118 DLGIEDGDVID 128
DLG+E GD+I+
Sbjct 125 DLGMESGDLIE 135
> ECU07g0660
Length=100
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 18/60 (30%), Positives = 34/60 (56%), Gaps = 0/60 (0%)
Query 63 LKVRSPDGSEVYFKIKRRTKLEKLMTAYCNRLGQSIDSVRFLYDGERVKPEKTPMDLGIE 122
L++ DG+ + F +K T +K++ A+ +G++ R L++G+ + KTP D G E
Sbjct 27 LRLVDQDGTTLVFNVKLSTTFKKILDAFSRNVGKNSSEFRILFNGKNIDLGKTPGDFGFE 86
> Hs8922069
Length=505
Score = 31.6 bits (70), Expect = 0.38, Method: Compositional matrix adjust.
Identities = 15/32 (46%), Positives = 21/32 (65%), Gaps = 0/32 (0%)
Query 16 HPFITFVFVEESESVSFIFFPFFLLISFLKMS 47
+P IT VF+E S S S+I F LL+ L++S
Sbjct 452 NPLITAVFLEASNSASYIQDAFQLLLPVLEIS 483
> SPBP8B7.23
Length=673
Score = 31.2 bits (69), Expect = 0.50, Method: Composition-based stats.
Identities = 16/60 (26%), Positives = 33/60 (55%), Gaps = 0/60 (0%)
Query 47 SEEKKDGENAEKEHMQLKVRSPDGSEVYFKIKRRTKLEKLMTAYCNRLGQSIDSVRFLYD 106
S ++K N+EK+ + ++ + K K + +LEK +A + +G+S+D + +YD
Sbjct 47 SADRKPHRNSEKKTQGMPRKNQQLASSERKTKNKKRLEKQSSAIADSIGESLDDPQTVYD 106
> SPBC365.11
Length=266
Score = 30.4 bits (67), Expect = 0.99, Method: Compositional matrix adjust.
Identities = 17/64 (26%), Positives = 34/64 (53%), Gaps = 1/64 (1%)
Query 51 KDGENAEKEHMQLKVRSPDGSEVYFKIKRRTK-LEKLMTAYCNRLGQSIDSVRFLYDGER 109
+D N ++EH+ +K R E K+K + L+ ++ N L +S D ++ L + ++
Sbjct 116 RDALNTKQEHLDIKKRLEKSDETVCKLKEENENLQDMLRNVGNELVESRDEIKELIEKQK 175
Query 110 VKPE 113
V+ E
Sbjct 176 VQKE 179
> Hs5453760
Length=81
Score = 29.6 bits (65), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 19/67 (28%), Positives = 33/67 (49%), Gaps = 0/67 (0%)
Query 61 MQLKVRSPDGSEVYFKIKRRTKLEKLMTAYCNRLGQSIDSVRFLYDGERVKPEKTPMDLG 120
M +KV++ G E+ I+ K+E++ + G R +Y G+++ EKT D
Sbjct 1 MLIKVKTLTGKEIEIDIEPTDKVERIKERVEEKEGIPPQQQRLIYSGKQMNDEKTAADYK 60
Query 121 IEDGDVI 127
I G V+
Sbjct 61 ILGGSVL 67
> 7292542
Length=1301
Score = 28.9 bits (63), Expect = 2.5, Method: Composition-based stats.
Identities = 15/65 (23%), Positives = 31/65 (47%), Gaps = 2/65 (3%)
Query 28 ESVSFI--FFPFFLLISFLKMSEEKKDGENAEKEHMQLKVRSPDGSEVYFKIKRRTKLEK 85
+ VSF+ + P +S + ++E+ + A ++H + +P SE Y I+R +
Sbjct 626 DCVSFLVSYLPLLQTLSQMPITEQVRRAALAWRQHKEQAQAAPGSSEAYHNIRREEVISN 685
Query 86 LMTAY 90
T +
Sbjct 686 ARTNW 690
> YDR363w
Length=456
Score = 28.9 bits (63), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 18/75 (24%), Positives = 37/75 (49%), Gaps = 3/75 (4%)
Query 59 EHMQLKVRSPDGSEVYFKIKRRTKLEKLMTAY--CNRLGQSIDSVRFLYDGERVKPEKTP 116
E M++ + D ++Y ++R T K+ Y +L Q V+ L+D + + +
Sbjct 382 EVMRIALMGQDNKKIYVHVRRSTPFSKIAEYYRIQKQLPQKT-RVKLLFDHDELDMNECI 440
Query 117 MDLGIEDGDVIDAMV 131
D +ED D++D ++
Sbjct 441 ADQDMEDEDMVDVII 455
> Hs19923899
Length=821
Score = 28.9 bits (63), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 17/53 (32%), Positives = 28/53 (52%), Gaps = 2/53 (3%)
Query 53 GENAEKEHMQLKVRSPDGSEVYFKIKR--RTKLEKLMTAYCNRLGQSIDSVRF 103
E +EH +LK R ++ K+KR R+K+EK + + + Q+ D V F
Sbjct 744 AEAISQEHQELKAREKSQAQTLHKVKRELRSKMEKEIQQLQDMITQNDDDVFF 796
> CE23468
Length=906
Score = 28.5 bits (62), Expect = 3.8, Method: Composition-based stats.
Identities = 11/38 (28%), Positives = 23/38 (60%), Gaps = 0/38 (0%)
Query 43 FLKMSEEKKDGENAEKEHMQLKVRSPDGSEVYFKIKRR 80
FL++ + ++D + E++ ++V +P S YF + RR
Sbjct 701 FLQLCDSREDQTPVDGENVNMRVANPTTSAQYFHLLRR 738
> 7303547
Length=1610
Score = 27.7 bits (60), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 17/63 (26%), Positives = 32/63 (50%), Gaps = 6/63 (9%)
Query 63 LKVRSPDGSEVYFKIKRRTKLEKLMTAYCNRLGQSIDSVRFLYDGERVKPEKTPMDLGIE 122
+K+R + + Y +I+RR K K Y ++ + +L + +R + E +D G +
Sbjct 800 IKLRWKNVRKRYVRIERRLKQGKRFKGYFDK------ATNYLENRDRPRSEWLAVDCGED 853
Query 123 DGD 125
DGD
Sbjct 854 DGD 856
> 7298505
Length=84
Score = 27.3 bits (59), Expect = 8.1, Method: Compositional matrix adjust.
Identities = 15/67 (22%), Positives = 34/67 (50%), Gaps = 0/67 (0%)
Query 61 MQLKVRSPDGSEVYFKIKRRTKLEKLMTAYCNRLGQSIDSVRFLYDGERVKPEKTPMDLG 120
M +KV++ G E+ I+ K++++ + G R ++ G+++ +KT D
Sbjct 1 MLIKVKTLTGKEIEIDIEPTDKVDRIKERVEEKEGIPPQQQRLIFSGKQMNDDKTAADYK 60
Query 121 IEDGDVI 127
++ G V+
Sbjct 61 VQGGSVL 67
Lambda K H
0.320 0.139 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1319765976
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40