bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0194_orf1
Length=128
Score E
Sequences producing significant alignments: (Bits) Value
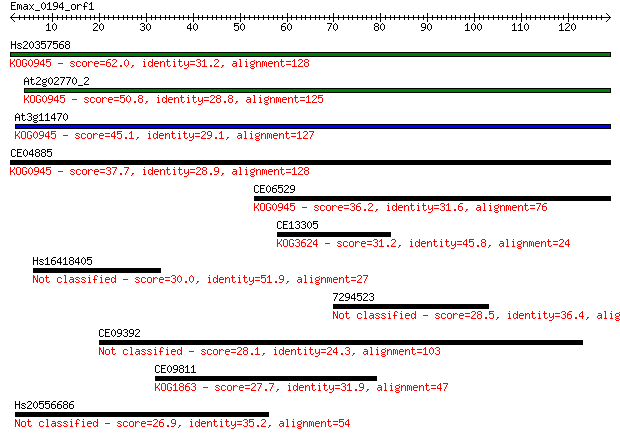
Hs20357568 62.0 3e-10
At2g02770_2 50.8 7e-07
At3g11470 45.1 3e-05
CE04885 37.7 0.005
CE06529 36.2 0.014
CE13305 31.2 0.53
Hs16418405 30.0 1.1
7294523 28.5 2.9
CE09392 28.1 4.2
CE09811 27.7 5.0
Hs20556686 26.9 9.1
> Hs20357568
Length=309
Score = 62.0 bits (149), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 40/130 (30%), Positives = 60/130 (46%), Gaps = 3/130 (2%)
Query 1 DRLRALSSRLLQRLLLSSYCCCNPQHIVIERKETRSKPIWKPSPGETADFVHFNVSHDEG 60
D A++ RL+ R L++ HI ++R + KP+ +FN+SH
Sbjct 56 DAKAAMAGRLMIRKLVAEKLNIPWNHIRLQRT-AKGKPVLAKDSSNPYPNFNFNISHQGD 114
Query 61 LVVFGASRHL-VGVDCMRCSPRGR-DISSFLQQMRRHCTLRDWSYIIAVHTPEQQLRRFM 118
V A L VG+D M+ S GR I F M+R T ++W I + QL F
Sbjct 115 YAVLAAEPELQVGIDIMKTSFPGRGSIPEFFHIMKRKFTNKEWETIRSFKDEWTQLDMFY 174
Query 119 RVWTVKEAFV 128
R W +KE+F+
Sbjct 175 RNWALKESFI 184
> At2g02770_2
Length=238
Score = 50.8 bits (120), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 36/131 (27%), Positives = 65/131 (49%), Gaps = 9/131 (6%)
Query 4 RALSSRLLQRLLLSSYCCCN---PQHIVIERKETRSKP--IWKPSPGETADFVHFNVSHD 58
AL +R L R ++ Y N P+ ++ +K KP W+ + +HFN+SH
Sbjct 68 NALLARTLVRTTIARYQTNNVVDPRTLIF-KKNMYGKPEVDWQSYKNCDSPPLHFNISHT 126
Query 59 EGLVVFGASRHL-VGVDCMRCSPRGRDISSFLQQMRRHCTLRDWSYIIAVHTPEQQLRRF 117
+ L+ G + H+ VG+D + + L R + + ++ A+ PE Q + F
Sbjct 127 DSLISCGVTVHVPVGIDLEEMERKIKH--DVLALAERFYSADEVKFLSAIPDPEVQRKEF 184
Query 118 MRVWTVKEAFV 128
+++WT+KEA+V
Sbjct 185 IKLWTLKEAYV 195
> At3g11470
Length=275
Score = 45.1 bits (105), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 37/135 (27%), Positives = 68/135 (50%), Gaps = 12/135 (8%)
Query 2 RLRALSSRLLQRLLLSSYCCCN---PQHIVIERKETRSKP--IWKPSPGETADFVHFNVS 56
+ AL +R L R ++ Y N P+ ++ +K KP W+ +HFN+S
Sbjct 66 KKNALLARTLVRTTIARYQTNNEVDPKALMF-KKNMYGKPEVDWQNYTNCNNPPLHFNIS 124
Query 57 HDEGLVVFGASRHL-VGVDCMRCSPRGR-DISSFLQQMRRHCTLRDWSYIIAVHTPEQQL 114
H + L+ G + H+ VG+D + + DI +F + R + + ++ + PE Q
Sbjct 125 HTDSLIACGVTVHVPVGIDVEDKERKIKHDILAFAE---RFYSADEVKFLSTLPDPEVQR 181
Query 115 RRFMRVWTVK-EAFV 128
+ F+++WT+K EA+V
Sbjct 182 KEFIKLWTLKVEAYV 196
> CE04885
Length=297
Score = 37.7 bits (86), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 37/131 (28%), Positives = 52/131 (39%), Gaps = 7/131 (5%)
Query 1 DRLRALSSRLLQRLLLSSYCCCNPQHIVIERKETRSKPIWKPSPGETADFVHFNVSHDEG 60
D L L RLL R + I ER E R KP P +T NVSH
Sbjct 52 DALACLFGRLLLRHSAQKFSGEPWNTIRFERTE-RGKPFL-AVPADTT--FGLNVSHQGD 107
Query 61 LVVFGAS-RHLVGVDCMRCSPR--GRDISSFLQQMRRHCTLRDWSYIIAVHTPEQQLRRF 117
V F +S VGVD MR + ++ M + + + + + T ++ F
Sbjct 108 YVAFASSCSSKVGVDVMRLDNERNNKTADEYINSMAKSASPEELRMMRSQPTEAMKMTMF 167
Query 118 MRVWTVKEAFV 128
R W +KEA +
Sbjct 168 YRYWCLKEAIL 178
> CE06529
Length=299
Score = 36.2 bits (82), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 24/80 (30%), Positives = 40/80 (50%), Gaps = 6/80 (7%)
Query 53 FNVSHDEGLVVFGASRHLVGVDCMRCSPRGRDISSF-LQQMRRHCTLRDWSYIIAVHTPE 111
+NVSH LVV +GVD MR + R+ +S + ++RH + + + E
Sbjct 97 YNVSHHGDLVVLATGDTRIGVDVMRVNEARRETASEQMNTLKRHFSENEIEMVKGGDKCE 156
Query 112 QQLRR---FMRVWTVKEAFV 128
L+R F R+W +KE+ +
Sbjct 157 --LKRWHAFYRIWCLKESIL 174
> CE13305
Length=732
Score = 31.2 bits (69), Expect = 0.53, Method: Composition-based stats.
Identities = 11/24 (45%), Positives = 16/24 (66%), Gaps = 0/24 (0%)
Query 58 DEGLVVFGASRHLVGVDCMRCSPR 81
D + +FG+SRHL+G +C PR
Sbjct 548 DLAMFLFGSSRHLIGSSFQKCLPR 571
> Hs16418405
Length=341
Score = 30.0 bits (66), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 14/27 (51%), Positives = 17/27 (62%), Gaps = 0/27 (0%)
Query 6 LSSRLLQRLLLSSYCCCNPQHIVIERK 32
LS R +Q LLL C +PQH V+ RK
Sbjct 150 LSPRYIQPLLLKEETCLDPQHPVMPRK 176
> 7294523
Length=226
Score = 28.5 bits (62), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 12/33 (36%), Positives = 16/33 (48%), Gaps = 0/33 (0%)
Query 70 LVGVDCMRCSPRGRDISSFLQQMRRHCTLRDWS 102
L C++C P + I FL R HC L +S
Sbjct 135 LAAAKCLQCIPLIKRIFEFLNDYRTHCELFSFS 167
> CE09392
Length=966
Score = 28.1 bits (61), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 25/103 (24%), Positives = 42/103 (40%), Gaps = 24/103 (23%)
Query 20 CCCNPQHIVIERKETRSKPIWKPSPGETADFVHFNVSHDEGLVVFGASRHLVGVDCMRCS 79
CC N I ++ + +W G V + E + V+ + +L+G MR S
Sbjct 503 CCANGTEIAVKGRVL----VWFKVKG---------VEYTEYVYVWNRNNNLLGTSWMRHS 549
Query 80 PRGRDISSFLQQMRRHCTLRDWSYIIAVHTPEQQLRRFMRVWT 122
P+ RD + + ++I V T +Q L F R+ T
Sbjct 550 PQMRDALAVM-----------VNHISTVDTSKQNLHSFSRIPT 581
> CE09811
Length=1230
Score = 27.7 bits (60), Expect = 5.0, Method: Composition-based stats.
Identities = 15/50 (30%), Positives = 26/50 (52%), Gaps = 3/50 (6%)
Query 32 KETRSKPIWKPSPGETA---DFVHFNVSHDEGLVVFGASRHLVGVDCMRC 78
KET KP +P+ + A + ++ V+ DE V F + + +DC +C
Sbjct 3 KETDKKPKERPNQRKVAFRNEAINALVNTDEDQVTFSKAMEVAKLDCQKC 52
> Hs20556686
Length=284
Score = 26.9 bits (58), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 19/54 (35%), Positives = 27/54 (50%), Gaps = 2/54 (3%)
Query 2 RLRALSSRLLQRLLLSSYCCCNPQHIVIERKETRSKPIWKPSPGETADFVHFNV 55
RLRALS+ L +++L P + ERK +R K S G +D H N+
Sbjct 124 RLRALSAAFLLQMILEGRRYFFPVGVYPERKSSRGKERESDSLG--SDMDHLNM 175
Lambda K H
0.328 0.138 0.452
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1213511838
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40