bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
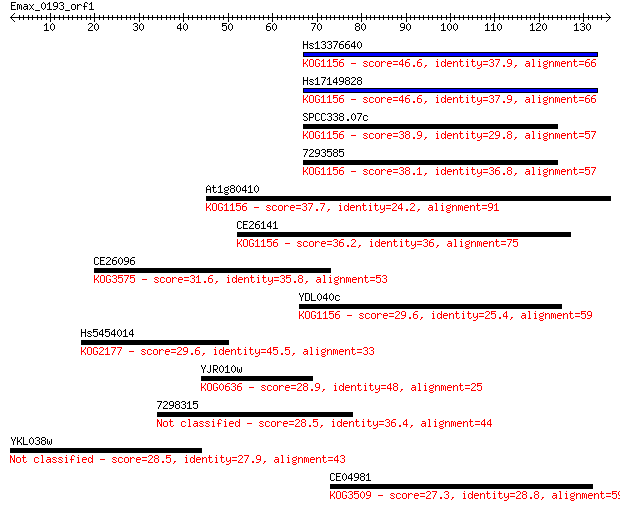
Query= Emax_0193_orf1
Length=135
Score E
Sequences producing significant alignments: (Bits) Value
Hs13376640 46.6 1e-05
Hs17149828 46.6 1e-05
SPCC338.07c 38.9 0.003
7293585 38.1 0.005
At1g80410 37.7 0.006
CE26141 36.2 0.017
CE26096 31.6 0.45
YDL040c 29.6 1.5
Hs5454014 29.6 1.7
YJR010w 28.9 2.6
7298315 28.5 3.3
YKL038w 28.5 3.6
CE04981 27.3 8.1
> Hs13376640
Length=526
Score = 46.6 bits (109), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 25/71 (35%), Positives = 39/71 (54%), Gaps = 5/71 (7%)
Query 67 LPLSFFTGDRFVAALDHFLRPMLRKGVVSIFAAIRRLY-TRNRVHLITSVIESYVTNLEK 125
LPL+F +G++F LD FLR KG +F +R LY + +V +I ++ Y T+L+
Sbjct 296 LPLNFLSGEKFKECLDKFLRMNFSKGCPPVFNTLRSLYKDKEKVAIIEELVVGYETSLKS 355
Query 126 ----KPSTFGK 132
P+ GK
Sbjct 356 CRLFNPNDDGK 366
> Hs17149828
Length=866
Score = 46.6 bits (109), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 25/71 (35%), Positives = 39/71 (54%), Gaps = 5/71 (7%)
Query 67 LPLSFFTGDRFVAALDHFLRPMLRKGVVSIFAAIRRLY-TRNRVHLITSVIESYVTNLEK 125
LPL+F +G++F LD FLR KG +F +R LY + +V +I ++ Y T+L+
Sbjct 296 LPLNFLSGEKFKECLDKFLRMNFSKGCPPVFNTLRSLYKDKEKVAIIEELVVGYETSLKS 355
Query 126 ----KPSTFGK 132
P+ GK
Sbjct 356 CRLFNPNDDGK 366
> SPCC338.07c
Length=729
Score = 38.9 bits (89), Expect = 0.003, Method: Composition-based stats.
Identities = 17/58 (29%), Positives = 33/58 (56%), Gaps = 1/58 (1%)
Query 67 LPLSFFTGDRFVAALDHFLRPMLRKGVVSIFAAIRRLYT-RNRVHLITSVIESYVTNL 123
LPL GD F+ +D +LR L++G+ S+F ++ LY + ++ ++ Y ++L
Sbjct 301 LPLEKLEGDEFLTHVDLYLRKKLKRGIPSVFVDVKSLYKDTKKCKVVEDLVSKYASSL 358
> 7293585
Length=890
Score = 38.1 bits (87), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 21/58 (36%), Positives = 29/58 (50%), Gaps = 1/58 (1%)
Query 67 LPLSFFTGDRFVAALDHFLRPMLRKGVVSIFAAIRRLYT-RNRVHLITSVIESYVTNL 123
LPL+ GD F D +LR LRKG+ +F +R L+ R +I + Y NL
Sbjct 296 LPLNIANGDEFRVVTDEYLRRGLRKGIPPLFVNVRTLHQIPERAAVIEELALQYFENL 353
> At1g80410
Length=683
Score = 37.7 bits (86), Expect = 0.006, Method: Composition-based stats.
Identities = 22/91 (24%), Positives = 42/91 (46%), Gaps = 3/91 (3%)
Query 45 ELIASYFEHLQRELAPFPLLDYLPLSFFTGDRFVAALDHFLRPMLRKGVVSIFAAIRRLY 104
E + + ++ L + + +PL F + F A+ +++P+L KGV S+F+ + LY
Sbjct 267 EKLNALYQSLSEQYTRSSAVKRIPLDFLQDENFKEAVAKYIKPLLTKGVPSLFSDLSSLY 326
Query 105 TRNRVHLITSVIESYVTNLEKKPSTFGKLLG 135
R ++E V ++ T G G
Sbjct 327 DHPRK---PDILEQLVVEMKHSIGTTGSFPG 354
> CE26141
Length=816
Score = 36.2 bits (82), Expect = 0.017, Method: Composition-based stats.
Identities = 27/75 (36%), Positives = 37/75 (49%), Gaps = 7/75 (9%)
Query 52 EHLQRELAPFPLLDYLPLSFFTGDRFVAALDHFLRPMLRKGVVSIFAAIRRLYTRNRVHL 111
E +R AP L YL G+ L ++ PMLRKG S+FA++ LY +
Sbjct 234 EKFKRAAAPRRLALYL----VEGEELRRRLHEWMIPMLRKGAPSLFASLVPLYKYPQKQ- 288
Query 112 ITSVIESYVTNLEKK 126
+VIES +T KK
Sbjct 289 --AVIESLITEYVKK 301
> CE26096
Length=374
Score = 31.6 bits (70), Expect = 0.45, Method: Compositional matrix adjust.
Identities = 19/59 (32%), Positives = 30/59 (50%), Gaps = 6/59 (10%)
Query 20 TKLYSKRTLTPHSIRVTREPTEREQELIASYFEHLQRELAPFP------LLDYLPLSFF 72
T ++SKR PH+ R T + RE +++ + + +LA FP LL + L FF
Sbjct 140 TSMFSKRIPKPHTFRDTDKLMFREYDMVTDWAMNSFPQLADFPNDQRKILLKHFYLQFF 198
> YDL040c
Length=854
Score = 29.6 bits (65), Expect = 1.5, Method: Composition-based stats.
Identities = 15/63 (23%), Positives = 33/63 (52%), Gaps = 4/63 (6%)
Query 66 YLPLSFFTG-DRFVAALDHFLRPMLRKGVVSIFAAIRRLYTRNRVH---LITSVIESYVT 121
++PL+F + L ++ P L +GV + F+ ++ LY R + L+ ++ Y++
Sbjct 313 FIPLTFLQDKEELSKKLREYVLPQLERGVPATFSNVKPLYQRRKSKVSPLLEKIVLDYLS 372
Query 122 NLE 124
L+
Sbjct 373 GLD 375
> Hs5454014
Length=465
Score = 29.6 bits (65), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 15/35 (42%), Positives = 20/35 (57%), Gaps = 2/35 (5%)
Query 17 CNITKLY--SKRTLTPHSIRVTREPTEREQELIAS 49
CN++KLY K+ L H + VT +P ELI S
Sbjct 274 CNVSKLYFDVKKMLRSHQVSVTLDPDTAHHELILS 308
> YJR010w
Length=511
Score = 28.9 bits (63), Expect = 2.6, Method: Composition-based stats.
Identities = 12/25 (48%), Positives = 16/25 (64%), Gaps = 0/25 (0%)
Query 44 QELIASYFEHLQRELAPFPLLDYLP 68
QEL+ SY L E+ PF ++ YLP
Sbjct 311 QELVESYKHELDIEVVPFRMVTYLP 335
> 7298315
Length=796
Score = 28.5 bits (62), Expect = 3.3, Method: Composition-based stats.
Identities = 16/45 (35%), Positives = 23/45 (51%), Gaps = 1/45 (2%)
Query 34 RVTREPTEREQELIASYFEHLQRELAPFPLLDYLPL-SFFTGDRF 77
R R+P + E S +EHL R PF LPL ++ + +RF
Sbjct 372 RYRRQPLDEEYHSFKSKYEHLSRRDNPFKGSSLLPLDTYPSENRF 416
> YKL038w
Length=1170
Score = 28.5 bits (62), Expect = 3.6, Method: Composition-based stats.
Identities = 12/43 (27%), Positives = 22/43 (51%), Gaps = 0/43 (0%)
Query 1 REERRVCTEAKQNGKECNITKLYSKRTLTPHSIRVTREPTERE 43
++E+ VC+ ++NG C+ ++ KR + R T P E
Sbjct 60 KDEKGVCSNCQRNGDRCSFDRVPLKRGPSKGYTRSTSHPRTNE 102
> CE04981
Length=757
Score = 27.3 bits (59), Expect = 8.1, Method: Composition-based stats.
Identities = 17/60 (28%), Positives = 29/60 (48%), Gaps = 1/60 (1%)
Query 73 TGDRFVAALDHFLRPMLRKGVVSIFAAIRRLYTRNRVHLITSVIESYVTNLEKKP-STFG 131
TG+ F+ +LD P + K + ++R+ + L+ S + VT + P STFG
Sbjct 510 TGNIFIGSLDDNDIPSVVKDIEGFAGCVKRIRLNGKAILMQSTYATNVTECFQDPCSTFG 569
Lambda K H
0.324 0.138 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1388795348
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40