bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0187_orf1
Length=205
Score E
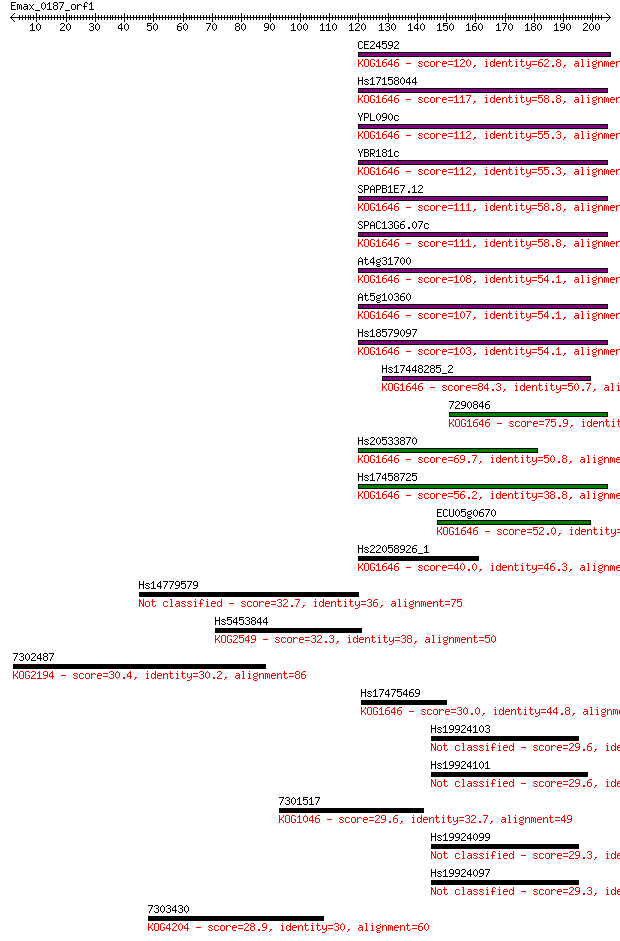
Sequences producing significant alignments: (Bits) Value
CE24592 120 2e-27
Hs17158044 117 1e-26
YPL090c 112 6e-25
YBR181c 112 6e-25
SPAPB1E7.12 111 9e-25
SPAC13G6.07c 111 9e-25
At4g31700 108 9e-24
At5g10360 107 1e-23
Hs18579097 103 3e-22
Hs17448285_2 84.3 1e-16
7290846 75.9 5e-14
Hs20533870 69.7 3e-12
Hs17458725 56.2 5e-08
ECU05g0670 52.0 9e-07
Hs22058926_1 40.0 0.004
Hs14779579 32.7 0.54
Hs5453844 32.3 0.76
7302487 30.4 2.3
Hs17475469 30.0 3.2
Hs19924103 29.6 4.2
Hs19924101 29.6 4.3
7301517 29.6 4.5
Hs19924099 29.3 5.9
Hs19924097 29.3 5.9
7303430 28.9 7.4
> CE24592
Length=246
Score = 120 bits (300), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 54/86 (62%), Positives = 66/86 (76%), Gaps = 0/86 (0%)
Query 120 MKLNIANPACGLQKTIEIDDEKKLLPFFERRIGMDIRGDSLGDEYKGYILRITGGNDKQG 179
M+LN A PA GLQK+ E+D+EKKL FFE+R+ ++ D+LGDE+KGY++RI GGNDKQG
Sbjct 1 MRLNFAYPATGLQKSFEVDEEKKLRLFFEKRMSQEVAIDALGDEWKGYVVRIGGGNDKQG 60
Query 180 FPMLQGILINNRVRLLFNNLMKCYRE 205
FPM QGIL N RVRLL CYRE
Sbjct 61 FPMKQGILTNGRVRLLLKKGQSCYRE 86
> Hs17158044
Length=249
Score = 117 bits (293), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 50/85 (58%), Positives = 67/85 (78%), Gaps = 0/85 (0%)
Query 120 MKLNIANPACGLQKTIEIDDEKKLLPFFERRIGMDIRGDSLGDEYKGYILRITGGNDKQG 179
MKLNI+ PA G QK IE+DDE+KL F+E+R+ ++ D+LG+E+KGY++RI+GGNDKQG
Sbjct 1 MKLNISFPATGCQKLIEVDDERKLRTFYEKRMATEVAADALGEEWKGYVVRISGGNDKQG 60
Query 180 FPMLQGILINNRVRLLFNNLMKCYR 204
FPM QG+L + RVRLL + CYR
Sbjct 61 FPMKQGVLTHGRVRLLLSKGHSCYR 85
> YPL090c
Length=236
Score = 112 bits (280), Expect = 6e-25, Method: Compositional matrix adjust.
Identities = 47/85 (55%), Positives = 66/85 (77%), Gaps = 0/85 (0%)
Query 120 MKLNIANPACGLQKTIEIDDEKKLLPFFERRIGMDIRGDSLGDEYKGYILRITGGNDKQG 179
MKLNI+ P G QKT EIDDE ++ FF++RIG ++ G+++GDE+KGY+ +I+GGNDKQG
Sbjct 1 MKLNISYPVNGSQKTFEIDDEHRIRVFFDKRIGQEVDGEAVGDEFKGYVFKISGGNDKQG 60
Query 180 FPMLQGILINNRVRLLFNNLMKCYR 204
FPM QG+L+ R++LL + CYR
Sbjct 61 FPMKQGVLLPTRIKLLLTKNVSCYR 85
> YBR181c
Length=236
Score = 112 bits (280), Expect = 6e-25, Method: Compositional matrix adjust.
Identities = 47/85 (55%), Positives = 66/85 (77%), Gaps = 0/85 (0%)
Query 120 MKLNIANPACGLQKTIEIDDEKKLLPFFERRIGMDIRGDSLGDEYKGYILRITGGNDKQG 179
MKLNI+ P G QKT EIDDE ++ FF++RIG ++ G+++GDE+KGY+ +I+GGNDKQG
Sbjct 1 MKLNISYPVNGSQKTFEIDDEHRIRVFFDKRIGQEVDGEAVGDEFKGYVFKISGGNDKQG 60
Query 180 FPMLQGILINNRVRLLFNNLMKCYR 204
FPM QG+L+ R++LL + CYR
Sbjct 61 FPMKQGVLLPTRIKLLLTKNVSCYR 85
> SPAPB1E7.12
Length=239
Score = 111 bits (278), Expect = 9e-25, Method: Compositional matrix adjust.
Identities = 50/85 (58%), Positives = 65/85 (76%), Gaps = 0/85 (0%)
Query 120 MKLNIANPACGLQKTIEIDDEKKLLPFFERRIGMDIRGDSLGDEYKGYILRITGGNDKQG 179
MKLNI+ PA G QK IEIDD+++L F E+R+G ++ GDS+G E+ GY+ +ITGGNDKQG
Sbjct 1 MKLNISYPANGTQKLIEIDDDRRLRVFMEKRMGQEVPGDSVGPEFAGYVFKITGGNDKQG 60
Query 180 FPMLQGILINNRVRLLFNNLMKCYR 204
FPM QG+L+ +RVRLL CYR
Sbjct 61 FPMFQGVLLPHRVRLLLRAGHPCYR 85
> SPAC13G6.07c
Length=239
Score = 111 bits (278), Expect = 9e-25, Method: Compositional matrix adjust.
Identities = 50/85 (58%), Positives = 65/85 (76%), Gaps = 0/85 (0%)
Query 120 MKLNIANPACGLQKTIEIDDEKKLLPFFERRIGMDIRGDSLGDEYKGYILRITGGNDKQG 179
MKLNI+ PA G QK IEIDD+++L F E+R+G ++ GDS+G E+ GY+ +ITGGNDKQG
Sbjct 1 MKLNISYPANGTQKLIEIDDDRRLRVFMEKRMGQEVPGDSVGPEFAGYVFKITGGNDKQG 60
Query 180 FPMLQGILINNRVRLLFNNLMKCYR 204
FPM QG+L+ +RVRLL CYR
Sbjct 61 FPMFQGVLLPHRVRLLLRAGHPCYR 85
> At4g31700
Length=250
Score = 108 bits (269), Expect = 9e-24, Method: Compositional matrix adjust.
Identities = 46/85 (54%), Positives = 63/85 (74%), Gaps = 0/85 (0%)
Query 120 MKLNIANPACGLQKTIEIDDEKKLLPFFERRIGMDIRGDSLGDEYKGYILRITGGNDKQG 179
MK N+ANP G QK +EIDD++KL F+++RI ++ GD+LG+E+KGY+ +I GG DKQG
Sbjct 1 MKFNVANPTTGCQKKLEIDDDQKLRAFYDKRISQEVSGDALGEEFKGYVFKIKGGCDKQG 60
Query 180 FPMLQGILINNRVRLLFNNLMKCYR 204
FPM QG+L RVRLL + C+R
Sbjct 61 FPMKQGVLTPGRVRLLLHRGTPCFR 85
> At5g10360
Length=249
Score = 107 bits (268), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 46/85 (54%), Positives = 63/85 (74%), Gaps = 0/85 (0%)
Query 120 MKLNIANPACGLQKTIEIDDEKKLLPFFERRIGMDIRGDSLGDEYKGYILRITGGNDKQG 179
MK N+ANP G QK +EIDD++KL FF++R+ ++ GD+LG+E+KGY+ +I GG DKQG
Sbjct 1 MKFNVANPTTGCQKKLEIDDDQKLRAFFDKRLSQEVSGDALGEEFKGYVFKIMGGCDKQG 60
Query 180 FPMLQGILINNRVRLLFNNLMKCYR 204
FPM QG+L RVRLL + C+R
Sbjct 61 FPMKQGVLTPGRVRLLLHRGTPCFR 85
> Hs18579097
Length=240
Score = 103 bits (256), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 46/85 (54%), Positives = 63/85 (74%), Gaps = 0/85 (0%)
Query 120 MKLNIANPACGLQKTIEIDDEKKLLPFFERRIGMDIRGDSLGDEYKGYILRITGGNDKQG 179
MKLNI+ P G QK IE+DDE KL F+E+ + ++ D+LG+E+KGY++RI+GGN+KQG
Sbjct 1 MKLNISFPVTGCQKLIEVDDECKLRTFYEKLMATEVAADTLGEEWKGYVVRISGGNNKQG 60
Query 180 FPMLQGILINNRVRLLFNNLMKCYR 204
FPM QG+L + RV LL + CYR
Sbjct 61 FPMKQGVLTHGRVHLLLSKGHSCYR 85
> Hs17448285_2
Length=155
Score = 84.3 bits (207), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 36/71 (50%), Positives = 54/71 (76%), Gaps = 0/71 (0%)
Query 128 ACGLQKTIEIDDEKKLLPFFERRIGMDIRGDSLGDEYKGYILRITGGNDKQGFPMLQGIL 187
A G QK IE+DD++KL F+E+ + ++ D+L +E+KGY++ I+GGNDKQ FPM +G+L
Sbjct 1 ATGCQKLIEVDDKRKLCTFYEKHMATEVAADALVEEWKGYVVLISGGNDKQWFPMKRGVL 60
Query 188 INNRVRLLFNN 198
I+ RVRLL +
Sbjct 61 IHGRVRLLMRD 71
> 7290846
Length=217
Score = 75.9 bits (185), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 33/54 (61%), Positives = 38/54 (70%), Gaps = 0/54 (0%)
Query 151 IGMDIRGDSLGDEYKGYILRITGGNDKQGFPMLQGILINNRVRLLFNNLMKCYR 204
+G + D LGDE+KGY LRI GGNDKQGFPM QG+L + RVRLL CYR
Sbjct 1 MGQVVEADILGDEWKGYQLRIAGGNDKQGFPMKQGVLTHGRVRLLLKKGHSCYR 54
> Hs20533870
Length=177
Score = 69.7 bits (169), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 31/61 (50%), Positives = 47/61 (77%), Gaps = 0/61 (0%)
Query 120 MKLNIANPACGLQKTIEIDDEKKLLPFFERRIGMDIRGDSLGDEYKGYILRITGGNDKQG 179
MKLN + A G QK I+++DE+KL F+E+ + +I +LG+E+KGY++RI+GGN+KQG
Sbjct 1 MKLNSSFLAIGCQKLIKVEDERKLRTFYEKHMATEIAAYALGEEWKGYVVRISGGNNKQG 60
Query 180 F 180
F
Sbjct 61 F 61
> Hs17458725
Length=248
Score = 56.2 bits (134), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 33/86 (38%), Positives = 45/86 (52%), Gaps = 19/86 (22%)
Query 120 MKLNI-ANPACGLQKTIEIDDEKKLLPFFERRIGMDIRGDSLGDEYKGYILRITGGNDKQ 178
MKLNI AC QK IE+D+E KL F+E+ + ++ D+LG E+KGY+
Sbjct 1 MKLNIFLATAC--QKLIEVDNEHKLHTFYEKCMATEVAADALGKEWKGYV---------- 48
Query 179 GFPMLQGILINNRVRLLFNNLMKCYR 204
G+L + V LL N CYR
Sbjct 49 ------GVLTHGHVHLLLNKRHACYR 68
> ECU05g0670
Length=203
Score = 52.0 bits (123), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 24/52 (46%), Positives = 35/52 (67%), Gaps = 0/52 (0%)
Query 147 FERRIGMDIRGDSLGDEYKGYILRITGGNDKQGFPMLQGILINNRVRLLFNN 198
++++IG G LG+E++G I+ ITGG+D QGFPM+ G L RVR L +
Sbjct 13 YDKKIGDQFDGGILGEEFEGTIMEITGGDDYQGFPMVSGHLTKKRVRPLLSK 64
> Hs22058926_1
Length=53
Score = 40.0 bits (92), Expect = 0.004, Method: Composition-based stats.
Identities = 19/41 (46%), Positives = 26/41 (63%), Gaps = 1/41 (2%)
Query 120 MKLNIANPACGLQKTIEIDDEKKLLPFFERRIGMDIRGDSL 160
MKLNI PA QK IE+DDE+KL +E + ++ D+L
Sbjct 1 MKLNIF-PAASCQKLIEVDDERKLRSLYEMPVATEVAADAL 40
> Hs14779579
Length=754
Score = 32.7 bits (73), Expect = 0.54, Method: Composition-based stats.
Identities = 27/79 (34%), Positives = 37/79 (46%), Gaps = 8/79 (10%)
Query 45 INLCILGDLYFFFPSFISLLTTYFLTSISLSRLNNLRILFALLGP-NLLGWWSTTTPPPT 103
++L +L DL P+ L L + R R + LGP + WSTT P P+
Sbjct 674 VDLAVLADL-VLTPARRGCLPCLLLKT---QRPQRHRTRPSALGPCSATSSWSTTMPWPS 729
Query 104 SPPS---PAAAAAAAGTRK 119
SP PAA A+A TR+
Sbjct 730 SPRPLRHPAAPGASARTRR 748
> Hs5453844
Length=622
Score = 32.3 bits (72), Expect = 0.76, Method: Composition-based stats.
Identities = 19/54 (35%), Positives = 27/54 (50%), Gaps = 4/54 (7%)
Query 71 SISLSRLNNLRILFALLGPNLLGWWSTTTPPPTSPPSPA----AAAAAAGTRKM 120
S+S++ + R L+A G +L + T P PT+P P AAA RKM
Sbjct 432 SLSVTLADIYRELYAFFGDSLATRFGTGQPAPTAPRPPGDKKEPAAAPDSVRKM 485
> 7302487
Length=687
Score = 30.4 bits (67), Expect = 2.3, Method: Composition-based stats.
Identities = 26/96 (27%), Positives = 50/96 (52%), Gaps = 10/96 (10%)
Query 2 LCLLSVLI----LCINSLYCLYCCLL--ILFCLINLCLYILFLL---ILVSVINLCILGD 52
+CL+ +++ L I S Y + C+L I+ ++NL L I+F+L + + +C +
Sbjct 432 ICLIVIMLTLTGLSIRSAYLIMLCILFDIVALIVNLVLIIIFILTAYLFAIAVTVCQILP 491
Query 53 LYFF-FPSFISLLTTYFLTSISLSRLNNLRILFALL 87
L FF + ++L+T + S S N ++ AL+
Sbjct 492 LVFFTYLCTVALVTLMPMQGRSGSSTNPDMVIAALV 527
> Hs17475469
Length=311
Score = 30.0 bits (66), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 13/29 (44%), Positives = 19/29 (65%), Gaps = 0/29 (0%)
Query 121 KLNIANPACGLQKTIEIDDEKKLLPFFER 149
K+ + PA G Q+ IE+DDE +L FE+
Sbjct 265 KMKLIFPATGCQELIEVDDEHELWTNFEK 293
> Hs19924103
Length=582
Score = 29.6 bits (65), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 18/52 (34%), Positives = 28/52 (53%), Gaps = 4/52 (7%)
Query 145 PFFERRIGMDIRGDSLGDEYKGYILRITGGNDK--QGFPMLQGILINNRVRL 194
PF + + DIR +G+ YK Y+ GN K G ML+ I +++R +L
Sbjct 306 PFIDSKY--DIRVQKIGNNYKAYMRTSISGNWKTNTGSAMLEQIAMSDRYKL 355
> Hs19924101
Length=478
Score = 29.6 bits (65), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 18/55 (32%), Positives = 29/55 (52%), Gaps = 4/55 (7%)
Query 145 PFFERRIGMDIRGDSLGDEYKGYILRITGGNDK--QGFPMLQGILINNRVRLLFN 197
PF + + DIR +G+ YK Y+ GN K G ML+ I +++R +L +
Sbjct 306 PFIDSKY--DIRVQKIGNNYKAYMRTSISGNWKTNTGSAMLEQIAMSDRYKLWVD 358
> 7301517
Length=1078
Score = 29.6 bits (65), Expect = 4.5, Method: Composition-based stats.
Identities = 16/51 (31%), Positives = 24/51 (47%), Gaps = 2/51 (3%)
Query 93 GWWSTTTPPP--TSPPSPAAAAAAAGTRKMKLNIANPACGLQKTIEIDDEK 141
GW S +PPP + P+P A+ T + +A PA + I + D K
Sbjct 96 GWVSMNSPPPLQAATPTPMASPTPTNTPTVTTTLAMPASSEKPEIRMVDPK 146
> Hs19924099
Length=705
Score = 29.3 bits (64), Expect = 5.9, Method: Composition-based stats.
Identities = 17/52 (32%), Positives = 27/52 (51%), Gaps = 4/52 (7%)
Query 145 PFFERRIGMDIRGDSLGDEYKGYILRITGGNDK--QGFPMLQGILINNRVRL 194
PF + + D+R +G YK Y+ GN K G ML+ I +++R +L
Sbjct 306 PFIDAK--YDVRVQKIGQNYKAYMRTSVSGNWKTNTGSAMLEQIAMSDRYKL 355
> Hs19924097
Length=669
Score = 29.3 bits (64), Expect = 5.9, Method: Composition-based stats.
Identities = 17/52 (32%), Positives = 27/52 (51%), Gaps = 4/52 (7%)
Query 145 PFFERRIGMDIRGDSLGDEYKGYILRITGGNDK--QGFPMLQGILINNRVRL 194
PF + + D+R +G YK Y+ GN K G ML+ I +++R +L
Sbjct 306 PFIDAK--YDVRVQKIGQNYKAYMRTSVSGNWKTNTGSAMLEQIAMSDRYKL 355
> 7303430
Length=2061
Score = 28.9 bits (63), Expect = 7.4, Method: Composition-based stats.
Identities = 18/72 (25%), Positives = 31/72 (43%), Gaps = 12/72 (16%)
Query 48 CILGDLYFF------------FPSFISLLTTYFLTSISLSRLNNLRILFALLGPNLLGWW 95
C + D FF + +F+ LT + +S + L L F + P+LL W+
Sbjct 916 CTISDAAFFDKVRKALRSPEVYDNFLRCLTLFNQEIVSKTELLGLVSPFLMKFPDLLRWF 975
Query 96 STTTPPPTSPPS 107
+ PP+ P+
Sbjct 976 TDFLGPPSGQPA 987
Lambda K H
0.331 0.148 0.468
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3658712052
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40