bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0179_orf1
Length=157
Score E
Sequences producing significant alignments: (Bits) Value
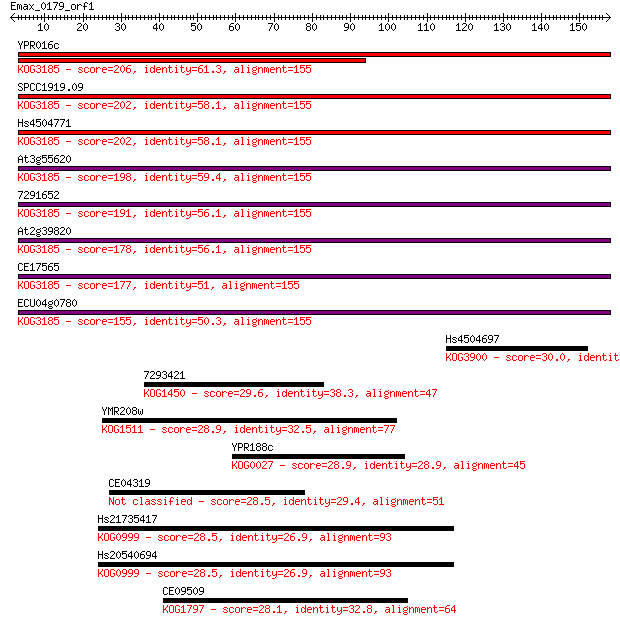
YPR016c 206 1e-53
SPCC1919.09 202 2e-52
Hs4504771 202 2e-52
At3g55620 198 4e-51
7291652 191 5e-49
At2g39820 178 5e-45
CE17565 177 5e-45
ECU04g0780 155 4e-38
Hs4504697 30.0 1.9
7293421 29.6 2.4
YMR208w 28.9 4.3
YPR188c 28.9 4.9
CE04319 28.5 5.9
Hs21735417 28.5 6.0
Hs20540694 28.5 6.0
CE09509 28.1 7.7
> YPR016c
Length=245
Score = 206 bits (524), Expect = 1e-53, Method: Compositional matrix adjust.
Identities = 95/155 (61%), Positives = 123/155 (79%), Gaps = 0/155 (0%)
Query 3 IPIIHTTISNTHVIGSLSVGNKHGLLLPIGTSDIEMNHIRNSLPEEIRIRRINDKLSALG 62
IPI+HTTI+ T +IG ++ GN+ GLL+P T+D E+ H+RNSLP+ ++I+R+ ++LSALG
Sbjct 46 IPIVHTTIAGTRIIGRMTAGNRRGLLVPTQTTDQELQHLRNSLPDSVKIQRVEERLSALG 105
Query 63 NCIVTNDYTALIHPDIDAETAEIIQDVLHVEVFRSIIGGQLLVGSYCVLTNQGGLVHSGT 122
N I NDY AL+HPDID ET E+I DVL VEVFR I G +LVGSYC L+NQGGLVH T
Sbjct 106 NVICCNDYVALVHPDIDRETEELISDVLGVEVFRQTISGNILVGSYCSLSNQGGLVHPQT 165
Query 123 SKEEMEDLSQLVQVPFTAGTVNRGSDLVGAGLMVN 157
S ++ E+LS L+QVP AGTVNRGS +VGAG++VN
Sbjct 166 SVQDQEELSSLLQVPLVAGTVNRGSSVVGAGMVVN 200
Score = 29.3 bits (64), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 24/92 (26%), Positives = 44/92 (47%), Gaps = 2/92 (2%)
Query 3 IPIIHTTISNTHVIGSL-SVGNKHGLLLPIGTSDIEMNHIRNSLPEEIRIRRINDKLSAL 61
+ + TIS ++GS S+ N+ GL+ P TS + + + L + +N S +
Sbjct 135 VEVFRQTISGNILVGSYCSLSNQGGLVHP-QTSVQDQEELSSLLQVPLVAGTVNRGSSVV 193
Query 62 GNCIVTNDYTALIHPDIDAETAEIIQDVLHVE 93
G +V NDY A+ D A +I+ + ++
Sbjct 194 GAGMVVNDYLAVTGLDTTAPELSVIESIFRLQ 225
> SPCC1919.09
Length=244
Score = 202 bits (514), Expect = 2e-52, Method: Compositional matrix adjust.
Identities = 90/155 (58%), Positives = 122/155 (78%), Gaps = 0/155 (0%)
Query 3 IPIIHTTISNTHVIGSLSVGNKHGLLLPIGTSDIEMNHIRNSLPEEIRIRRINDKLSALG 62
+P++HTTI T +IG L+ GN+ GLL+P T+D E+ H+RNSLP+ ++I+R++++LSALG
Sbjct 46 VPVVHTTIGGTRIIGRLTCGNRKGLLVPSSTTDNELQHLRNSLPDPVKIQRVDERLSALG 105
Query 63 NCIVTNDYTALIHPDIDAETAEIIQDVLHVEVFRSIIGGQLLVGSYCVLTNQGGLVHSGT 122
N + NDY AL+HPDI+ ET EII DVL VEVFR + G +L GSYC L+NQG LVH T
Sbjct 106 NIVACNDYVALVHPDIERETEEIIADVLDVEVFRQTVAGNVLTGSYCALSNQGALVHPRT 165
Query 123 SKEEMEDLSQLVQVPFTAGTVNRGSDLVGAGLMVN 157
S +E ++LS L+QVP AGT+NRGSD++GAGL+VN
Sbjct 166 SIQEQDELSSLLQVPLVAGTINRGSDVIGAGLVVN 200
> Hs4504771
Length=245
Score = 202 bits (514), Expect = 2e-52, Method: Compositional matrix adjust.
Identities = 90/155 (58%), Positives = 121/155 (78%), Gaps = 0/155 (0%)
Query 3 IPIIHTTISNTHVIGSLSVGNKHGLLLPIGTSDIEMNHIRNSLPEEIRIRRINDKLSALG 62
IP++H +I+ +IG + VGN+HGLL+P T+D E+ HIRNSLP+ ++IRR+ ++LSALG
Sbjct 46 IPVVHASIAGCRIIGRMCVGNRHGLLVPNNTTDQELQHIRNSLPDTVQIRRVEERLSALG 105
Query 63 NCIVTNDYTALIHPDIDAETAEIIQDVLHVEVFRSIIGGQLLVGSYCVLTNQGGLVHSGT 122
N NDY AL+HPD+D ET EI+ DVL VEVFR + Q+LVGSYCV +NQGGLVH T
Sbjct 106 NVTTCNDYVALVHPDLDRETEEILADVLKVEVFRQTVADQVLVGSYCVFSNQGGLVHPKT 165
Query 123 SKEEMEDLSQLVQVPFTAGTVNRGSDLVGAGLMVN 157
S E+ ++LS L+QVP AGTVNRGS+++ AG++VN
Sbjct 166 SIEDQDELSSLLQVPLVAGTVNRGSEVIAAGMVVN 200
> At3g55620
Length=245
Score = 198 bits (504), Expect = 4e-51, Method: Compositional matrix adjust.
Identities = 92/155 (59%), Positives = 121/155 (78%), Gaps = 0/155 (0%)
Query 3 IPIIHTTISNTHVIGSLSVGNKHGLLLPIGTSDIEMNHIRNSLPEEIRIRRINDKLSALG 62
IPI+ T+I T +IG L GNK+GLL+P T+D E+ H+RNSLP+++ ++RI+++LSALG
Sbjct 46 IPIVKTSIGGTRIIGRLCAGNKNGLLVPHTTTDQELQHLRNSLPDQVVVQRIDERLSALG 105
Query 63 NCIVTNDYTALIHPDIDAETAEIIQDVLHVEVFRSIIGGQLLVGSYCVLTNQGGLVHSGT 122
NCI NDY AL H D+D ET EII DVL VEVFR I G +LVGSYC L+N+GG+VH T
Sbjct 106 NCIACNDYVALAHTDLDKETEEIIADVLGVEVFRQTIAGNILVGSYCALSNKGGMVHPHT 165
Query 123 SKEEMEDLSQLVQVPFTAGTVNRGSDLVGAGLMVN 157
S E++E+LS L+QVP AGTVNRGS+++ AG+ VN
Sbjct 166 SVEDLEELSTLLQVPLVAGTVNRGSEVIAAGMTVN 200
> 7291652
Length=245
Score = 191 bits (485), Expect = 5e-49, Method: Compositional matrix adjust.
Identities = 87/155 (56%), Positives = 118/155 (76%), Gaps = 0/155 (0%)
Query 3 IPIIHTTISNTHVIGSLSVGNKHGLLLPIGTSDIEMNHIRNSLPEEIRIRRINDKLSALG 62
IP++H + +IG L+VGN++GLL+P T+D E+ H+RNSLP+ ++I R+ ++LSALG
Sbjct 46 IPVVHANVGGCRIIGRLTVGNRNGLLVPNSTTDEELQHLRNSLPDAVKIYRVEERLSALG 105
Query 63 NCIVTNDYTALIHPDIDAETAEIIQDVLHVEVFRSIIGGQLLVGSYCVLTNQGGLVHSGT 122
N I NDY AL+HPD+D ET EII DVL VEVFR I LVGSY VL+NQGG+VH T
Sbjct 106 NVIACNDYVALVHPDLDKETEEIIADVLKVEVFRQTIADNSLVGSYAVLSNQGGMVHPKT 165
Query 123 SKEEMEDLSQLVQVPFTAGTVNRGSDLVGAGLMVN 157
S ++ ++LS L+QVP AGTVNRGS+++ AG++VN
Sbjct 166 SIQDQDELSSLLQVPLVAGTVNRGSEVLAAGMVVN 200
> At2g39820
Length=247
Score = 178 bits (451), Expect = 5e-45, Method: Compositional matrix adjust.
Identities = 87/155 (56%), Positives = 113/155 (72%), Gaps = 0/155 (0%)
Query 3 IPIIHTTISNTHVIGSLSVGNKHGLLLPIGTSDIEMNHIRNSLPEEIRIRRINDKLSALG 62
IPI+ T+I + IGSL VGNK+GLLL +D E+ H+R+SLP+E+ ++RI + + ALG
Sbjct 48 IPIVTTSIGGSGTIGSLCVGNKNGLLLSHTITDQELQHLRDSLPDEVVVQRIEEPICALG 107
Query 63 NCIVTNDYTALIHPDIDAETAEIIQDVLHVEVFRSIIGGQLLVGSYCVLTNQGGLVHSGT 122
N I NDY AL+HP ++ +T EII DVL VEV+R I LVGSYC L+N GG+VHS T
Sbjct 108 NAIACNDYVALVHPKLEKDTEEIISDVLGVEVYRQTIANNELVGSYCSLSNNGGMVHSNT 167
Query 123 SKEEMEDLSQLVQVPFTAGTVNRGSDLVGAGLMVN 157
+ EEM +L+ LVQVP AGTVNRGS ++ AGL VN
Sbjct 168 NVEEMVELANLVQVPLVAGTVNRGSQVISAGLTVN 202
> CE17565
Length=246
Score = 177 bits (450), Expect = 5e-45, Method: Compositional matrix adjust.
Identities = 79/155 (50%), Positives = 119/155 (76%), Gaps = 0/155 (0%)
Query 3 IPIIHTTISNTHVIGSLSVGNKHGLLLPIGTSDIEMNHIRNSLPEEIRIRRINDKLSALG 62
IP++HT+I++T ++G L+VGN+HGLL+P T+D E+ H+RNSLP+E+ IRR++++LSALG
Sbjct 46 IPVVHTSIASTRIVGRLTVGNRHGLLVPNATTDQELQHLRNSLPDEVAIRRVDERLSALG 105
Query 63 NCIVTNDYTALIHPDIDAETAEIIQDVLHVEVFRSIIGGQLLVGSYCVLTNQGGLVHSGT 122
N I ND+ A++H +I AET + + +VL VEVFR + LVGSYC+L++ G LV + T
Sbjct 106 NVIACNDHVAIVHAEISAETEQALVEVLKVEVFRVSLAQNSLVGSYCILSSNGCLVAART 165
Query 123 SKEEMEDLSQLVQVPFTAGTVNRGSDLVGAGLMVN 157
E +++ L+Q+P AGT NRGS+L+GAG++VN
Sbjct 166 PPETQREIAALLQIPVVAGTCNRGSELIGAGMVVN 200
> ECU04g0780
Length=248
Score = 155 bits (391), Expect = 4e-38, Method: Compositional matrix adjust.
Identities = 78/155 (50%), Positives = 112/155 (72%), Gaps = 0/155 (0%)
Query 3 IPIIHTTISNTHVIGSLSVGNKHGLLLPIGTSDIEMNHIRNSLPEEIRIRRINDKLSALG 62
IPI+ TTI++ +GS GN+HGLL+P +D E+ HIRNSLPE++ +RRI ++L+ALG
Sbjct 51 IPIVETTINSIRSVGSQCRGNRHGLLVPHTITDQEIMHIRNSLPEDVVVRRIEERLNALG 110
Query 63 NCIVTNDYTALIHPDIDAETAEIIQDVLHVEVFRSIIGGQLLVGSYCVLTNQGGLVHSGT 122
N I+ ND+ A+IH D+D E+ ++I+DVL V V+R IG + LVG++ L NQG LVH T
Sbjct 111 NVILCNDHIAIIHGDLDKESEDLIRDVLQVHVYRQNIGQEPLVGTFGALNNQGMLVHPFT 170
Query 123 SKEEMEDLSQLVQVPFTAGTVNRGSDLVGAGLMVN 157
S E ++LS+L++V AGT+N GS VG G++ N
Sbjct 171 STECQKELSELLEVNVVAGTINGGSQCVGGGVVAN 205
> Hs4504697
Length=366
Score = 30.0 bits (66), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 14/37 (37%), Positives = 18/37 (48%), Gaps = 0/37 (0%)
Query 115 GGLVHSGTSKEEMEDLSQLVQVPFTAGTVNRGSDLVG 151
GG H G+ EE ED+SQ + P T + S G
Sbjct 65 GGFTHRGSEPEEEEDVSQAILFPATDASCEDKSAARG 101
> 7293421
Length=581
Score = 29.6 bits (65), Expect = 2.4, Method: Composition-based stats.
Identities = 18/49 (36%), Positives = 25/49 (51%), Gaps = 2/49 (4%)
Query 36 IEMNHIRNSLPEEIRIRRIND--KLSALGNCIVTNDYTALIHPDIDAET 82
IE + R++ P E R+ D K+ L I NDY +L PD+D T
Sbjct 316 IEQKYRRSTQPIEGIYRQCGDYNKIQTLRFSIDANDYDSLRQPDVDIHT 364
> YMR208w
Length=443
Score = 28.9 bits (63), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 25/81 (30%), Positives = 38/81 (46%), Gaps = 10/81 (12%)
Query 25 HGLLLPIGTSDIEMNHIRNSLPEEIRIRRINDKLSALGNCIVTNDYTALIHPDIDAET-- 82
HGLL+ IG S + I+N L +++RI + G C +T L+ DI E
Sbjct 319 HGLLVSIGVSHPGLELIKN-LSDDLRIGSTKLTGAGGGGCSLT-----LLRRDITQEQID 372
Query 83 --AEIIQDVLHVEVFRSIIGG 101
+ +QD E F + +GG
Sbjct 373 SFKKKLQDDFSYETFETDLGG 393
> YPR188c
Length=163
Score = 28.9 bits (63), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 13/45 (28%), Positives = 26/45 (57%), Gaps = 1/45 (2%)
Query 59 SALGNCIVTNDYTALIHPDIDAETAEIIQDVLHVEVFRSIIGGQL 103
+ LG + +++ ++ PD D TAE+ ++ + +F SI+G L
Sbjct 45 ATLGKTLTDEEWSKMV-PDNDTSTAEVGEEGVSFPIFLSIMGKNL 88
> CE04319
Length=665
Score = 28.5 bits (62), Expect = 5.9, Method: Composition-based stats.
Identities = 15/51 (29%), Positives = 27/51 (52%), Gaps = 0/51 (0%)
Query 27 LLLPIGTSDIEMNHIRNSLPEEIRIRRINDKLSALGNCIVTNDYTALIHPD 77
LLLPI +S ++ + I + RI+ + ++ + + I +DY HPD
Sbjct 363 LLLPIVSSFLDNSPISEVEKSQARIKNQKNMIAMMEDLIEVHDYNMKNHPD 413
> Hs21735417
Length=824
Score = 28.5 bits (62), Expect = 6.0, Method: Composition-based stats.
Identities = 25/94 (26%), Positives = 44/94 (46%), Gaps = 6/94 (6%)
Query 24 KHGLLLPIGTSDIEMNHIRNSLPEEI-RIRRINDKLSALGNCIVTNDYTALIHPDIDAET 82
K GLL + + ++ H R SL E+ ++ R+ + LSAL + + + + D ++
Sbjct 359 KAGLLATLQDTQKQLEHTRGSLSEQQEKVTRLTENLSALRRLQASKERQTALDNEKDRDS 418
Query 83 AEIIQDVLHVEVFRSIIGGQLLVGSYCVLTNQGG 116
E D + EV I G ++L Y V + G
Sbjct 419 HE---DGDYYEV--DINGPEILACKYHVAVAEAG 447
> Hs20540694
Length=855
Score = 28.5 bits (62), Expect = 6.0, Method: Composition-based stats.
Identities = 25/94 (26%), Positives = 44/94 (46%), Gaps = 6/94 (6%)
Query 24 KHGLLLPIGTSDIEMNHIRNSLPEEI-RIRRINDKLSALGNCIVTNDYTALIHPDIDAET 82
K GLL + + ++ H R SL E+ ++ R+ + LSAL + + + + D ++
Sbjct 359 KAGLLATLQDTQKQLEHTRGSLSEQQEKVTRLTENLSALRRLQASKERQTALDNEKDRDS 418
Query 83 AEIIQDVLHVEVFRSIIGGQLLVGSYCVLTNQGG 116
E D + EV I G ++L Y V + G
Sbjct 419 HE---DGDYYEV--DINGPEILACKYHVAVAEAG 447
> CE09509
Length=1576
Score = 28.1 bits (61), Expect = 7.7, Method: Composition-based stats.
Identities = 21/64 (32%), Positives = 31/64 (48%), Gaps = 1/64 (1%)
Query 41 IRNSLPEEIRIRRINDKLSALGNCIVTNDYTALIHPDIDAETAEIIQDVLHVEVFRSIIG 100
I S+ E RR+ KLS GNC +D+ +L DI I +D++ E I
Sbjct 846 IFESMKEPDGARRVLIKLSRSGNCSNFDDWMSL-RDDIYDMANGIYEDLVTTEEALEIFA 904
Query 101 GQLL 104
G++L
Sbjct 905 GEIL 908
Lambda K H
0.318 0.137 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2106641548
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40