bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0127_orf1
Length=176
Score E
Sequences producing significant alignments: (Bits) Value
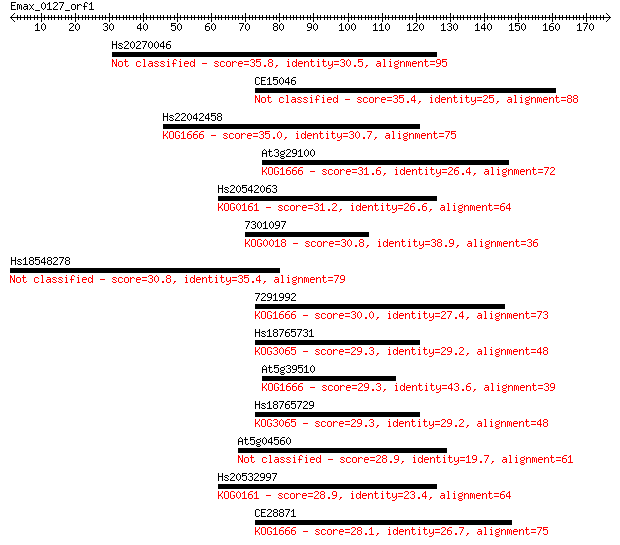
Hs20270046 35.8 0.040
CE15046 35.4 0.066
Hs22042458 35.0 0.079
At3g29100 31.6 0.88
Hs20542063 31.2 0.99
7301097 30.8 1.4
Hs18548278 30.8 1.6
7291992 30.0 2.6
Hs18765731 29.3 3.7
At5g39510 29.3 3.8
Hs18765729 29.3 4.5
At5g04560 28.9 5.1
Hs20532997 28.9 5.9
CE28871 28.1 8.6
> Hs20270046
Length=4169
Score = 35.8 bits (81), Expect = 0.040, Method: Composition-based stats.
Identities = 29/102 (28%), Positives = 47/102 (46%), Gaps = 10/102 (9%)
Query 31 CSWASSGPSGVAGVIFIPLGFLICPCVSTHALQLSLARTQQQAAQTEEMGG-------AI 83
CSW S+ P + V + L F P VS ++ +R ++ +++E+ GG A
Sbjct 2427 CSWDSNLPESLESVSDVLLNFF--PYVSPKT-SITDSREEEGVSESEDGGGSSVDSLAAH 2483
Query 84 LQRLQQQNEQLDGVKDGLDNVEMNIGRTKKTAHAIAKNAAHD 125
++ L Q L+ K+ L N E R + A + N AHD
Sbjct 2484 VKNLLQCESSLNHAKEILRNAEEEESRVRAHAWNMKFNLAHD 2525
> CE15046
Length=363
Score = 35.4 bits (80), Expect = 0.066, Method: Compositional matrix adjust.
Identities = 22/90 (24%), Positives = 43/90 (47%), Gaps = 7/90 (7%)
Query 73 AAQTEEMGGAILQRLQQQNEQLDGVKDGLDNVEMNIGRTKKTAHAIAKNAA--HDRCLQC 130
A +T+++ GA L + +QL + +G+D VE + T++ A+ + A + +
Sbjct 25 AGKTKDLAGAQLTK-----DQLGEIIEGIDQVESKVATTERAMEALTEAADLLDNYVTET 79
Query 131 LCCCVVLLLIASIVLIAQAVSSALHVCISR 160
C + I I++ AQ + LH + R
Sbjct 80 QKCTEKIDEIVEIIVAAQDIRDVLHATLKR 109
> Hs22042458
Length=175
Score = 35.0 bits (79), Expect = 0.079, Method: Compositional matrix adjust.
Identities = 23/78 (29%), Positives = 44/78 (56%), Gaps = 5/78 (6%)
Query 46 FIPLGFLIC-PCVST-HALQLSLARTQQQ-AAQTEEMGGAILQRLQQQNEQLDGVKDGLD 102
+ PL F C P +S Q LA+ ++ A +T+++G I++ L +Q +QL+ K L
Sbjct 68 YAPLSF--CNPMMSKLRNYQKDLAKLHREIATETDQIGSEIIEELGEQRDQLERTKSRLV 125
Query 103 NVEMNIGRTKKTAHAIAK 120
N N+G ++K ++++
Sbjct 126 NTSENLGNSRKILRSMSR 143
> At3g29100
Length=221
Score = 31.6 bits (70), Expect = 0.88, Method: Compositional matrix adjust.
Identities = 19/72 (26%), Positives = 38/72 (52%), Gaps = 3/72 (4%)
Query 75 QTEEMGGAILQRLQQQNEQLDGVKDGLDNVEMNIGRTKKTAHAIAKNAAHDRCLQCLCCC 134
+TEE+G +ILQ L Q + L + L V+ N+G++KK + + ++
Sbjct 146 ETEELGVSILQDLHGQRQSLLRAHETLHGVDDNVGKSKKILTTMTRRMNRNK---WTIGA 202
Query 135 VVLLLIASIVLI 146
++ +L+ +I+ I
Sbjct 203 IITVLVLAIIFI 214
> Hs20542063
Length=1939
Score = 31.2 bits (69), Expect = 0.99, Method: Composition-based stats.
Identities = 17/65 (26%), Positives = 31/65 (47%), Gaps = 1/65 (1%)
Query 62 LQLSLARTQQQAAQTEEMGGAILQRLQQQNEQLDGVKDGLDNVEMNIGRTKKTAHAI-AK 120
LQL + ++QA + EE L + ++ +LD ++ D E + + + + I AK
Sbjct 1873 LQLKVKAYKRQAEEAEEQANTNLSKFRKVQHELDEAEERADIAESQVNKLRAKSRDIGAK 1932
Query 121 NAAHD 125
HD
Sbjct 1933 QKMHD 1937
> 7301097
Length=1238
Score = 30.8 bits (68), Expect = 1.4, Method: Composition-based stats.
Identities = 14/39 (35%), Positives = 24/39 (61%), Gaps = 3/39 (7%)
Query 70 QQQAAQTEEMGGAILQ---RLQQQNEQLDGVKDGLDNVE 105
QQ +Q +E G IL+ R+Q + E + +K+ ++NVE
Sbjct 760 QQVQSQLDEFGPKILEIERRMQNREEHIQEIKENMNNVE 798
> Hs18548278
Length=250
Score = 30.8 bits (68), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 28/101 (27%), Positives = 39/101 (38%), Gaps = 22/101 (21%)
Query 1 VCCRRLSPVPQAPMFPCVSSFVSIDCHLFPC-SWASS---------GPSGVAGVIFIPLG 50
C R P Q+ + S F++ID LFP +W S+ P IF PL
Sbjct 63 TSCSRFQPGKQSQLLTHASGFLTIDLFLFPPDTWCSALVIVLRGHEYPPLKLPEIFFPLK 122
Query 51 FLICPCVS------------THALQLSLARTQQQAAQTEEM 79
L P S +H L +LA + + + TE M
Sbjct 123 KLDSPTESASTGAVKDTMYVSHGLTGNLAPAKPEQSATEAM 163
> 7291992
Length=230
Score = 30.0 bits (66), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 20/74 (27%), Positives = 35/74 (47%), Gaps = 1/74 (1%)
Query 73 AAQTEEMGGAILQRLQQQNEQLDGVKDGLDNVEMNIGRTKKTAHAIAKNAAHDR-CLQCL 131
A +TE++G +L L Q E L G + L +GR +T + + A ++ L +
Sbjct 145 ALETEQLGAQVLNDLHHQRETLQGARARLRETNAELGRASRTLNTMMLRALREKVVLYGV 204
Query 132 CCCVVLLLIASIVL 145
C V+ + S+ L
Sbjct 205 GVCFVVAVGVSLYL 218
> Hs18765731
Length=158
Score = 29.3 bits (64), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 14/48 (29%), Positives = 29/48 (60%), Gaps = 0/48 (0%)
Query 73 AAQTEEMGGAILQRLQQQNEQLDGVKDGLDNVEMNIGRTKKTAHAIAK 120
A ++++ G + L +Q EQL+ +++GLD + ++ T+KT + K
Sbjct 31 AIESQDAGIKTITMLDEQKEQLNRIEEGLDQINKDMRETEKTLTELNK 78
> At5g39510
Length=221
Score = 29.3 bits (64), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 17/39 (43%), Positives = 25/39 (64%), Gaps = 0/39 (0%)
Query 75 QTEEMGGAILQRLQQQNEQLDGVKDGLDNVEMNIGRTKK 113
+TEE+G +ILQ L Q + L + L V+ NIG++KK
Sbjct 146 ETEEIGVSILQDLHGQRQSLLRAHETLHGVDDNIGKSKK 184
> Hs18765729
Length=211
Score = 29.3 bits (64), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 14/48 (29%), Positives = 29/48 (60%), Gaps = 0/48 (0%)
Query 73 AAQTEEMGGAILQRLQQQNEQLDGVKDGLDNVEMNIGRTKKTAHAIAK 120
A ++++ G + L +Q EQL+ +++GLD + ++ T+KT + K
Sbjct 31 AIESQDAGIKTITMLDEQKEQLNRIEEGLDQINKDMRETEKTLTELNK 78
> At5g04560
Length=1017
Score = 28.9 bits (63), Expect = 5.1, Method: Composition-based stats.
Identities = 12/61 (19%), Positives = 31/61 (50%), Gaps = 0/61 (0%)
Query 68 RTQQQAAQTEEMGGAILQRLQQQNEQLDGVKDGLDNVEMNIGRTKKTAHAIAKNAAHDRC 127
+T+ Q E + G + ++ +++ DG + G + ++ G +K + + K A ++C
Sbjct 755 KTKIQKVVQENLHGMPPEVIEIEDDPTDGARKGKNTASISKGASKGNSSPVKKTAEKEKC 814
Query 128 L 128
+
Sbjct 815 I 815
> Hs20532997
Length=975
Score = 28.9 bits (63), Expect = 5.9, Method: Composition-based stats.
Identities = 15/64 (23%), Positives = 27/64 (42%), Gaps = 0/64 (0%)
Query 62 LQLSLARTQQQAAQTEEMGGAILQRLQQQNEQLDGVKDGLDNVEMNIGRTKKTAHAIAKN 121
LQL + +QQ E L + ++Q +L+ VK+ + E + + K A K
Sbjct 912 LQLKVQNYKQQVEVAETQANQYLSKYKKQQHELNEVKERAEVAESQVNKLKIKAREFGKK 971
Query 122 AAHD 125
+
Sbjct 972 VQEE 975
> CE28871
Length=224
Score = 28.1 bits (61), Expect = 8.6, Method: Compositional matrix adjust.
Identities = 20/75 (26%), Positives = 36/75 (48%), Gaps = 0/75 (0%)
Query 73 AAQTEEMGGAILQRLQQQNEQLDGVKDGLDNVEMNIGRTKKTAHAIAKNAAHDRCLQCLC 132
A +TE++G +L L Q E + +D L ++GR KT ++ + A +R L +
Sbjct 149 AVETEQIGAEMLSNLASQRETIGRSRDRLRQSNADLGRANKTLSSMIRRAIQNRLLLLIV 208
Query 133 CCVVLLLIASIVLIA 147
++ + IV A
Sbjct 209 TFLLSFMFLYIVYKA 223
Lambda K H
0.328 0.137 0.434
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2707167450
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40