bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
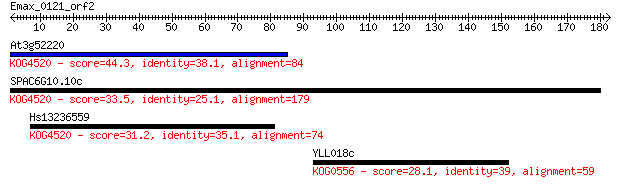
Query= Emax_0121_orf2
Length=182
Score E
Sequences producing significant alignments: (Bits) Value
At3g52220 44.3 1e-04
SPAC6G10.10c 33.5 0.23
Hs13236559 31.2 1.2
YLL018c 28.1 9.5
> At3g52220
Length=237
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 32/85 (37%), Positives = 46/85 (54%), Gaps = 4/85 (4%)
Query 1 GVRGGWEDFKWESLKTQTNADRDYYLGASAKVGICTRGGKFEKYDWWTKKKEADGPS-GA 59
GVRGG + F W+ +K R+ YLG S K + R K + W+ + K+ G A
Sbjct 8 GVRGGRDQFSWDEVKADKY--RENYLGHSIKAPVG-RWQKGKDLHWYARDKKQKGSEMDA 64
Query 60 EDEELRRVKRFEQQLLEEALGRRPR 84
EE++RVK E+Q + EALG P+
Sbjct 65 MKEEIQRVKEQEEQAMREALGLAPK 89
> SPAC6G10.10c
Length=194
Score = 33.5 bits (75), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 45/186 (24%), Positives = 71/186 (38%), Gaps = 45/186 (24%)
Query 1 GVRGGWEDFKWESLKTQTNADRDYYLGASAKVGICTRGGKFEKYDWWTKKKEADGPSGAE 60
G RGG F+WE ++ R YLG S R + + +WW+K + +E
Sbjct 7 GTRGGQAQFQWEEVRNDKQKGR--YLGQSI-YAASGRWAEGKDLEWWSKGRTTQSAINSE 63
Query 61 D-------EELRRVKRFEQQLLEEALGRRPRHLLAGEALADEEPAEPPVATTAAADDRRA 113
+ +E+ +K EQ++L EALG P + A +A
Sbjct 64 NSDKEKYKKEILEIKEREQRMLAEALGL-------------------PQPSALALTSSKA 104
Query 114 AEKALKKLKKEQRKLKKLRKKEHKREAKRLRLKRERHSESRSPRRRSSSVDSERRGDSHR 173
A ++ +K+ R + H R RH RR+ + ER +HR
Sbjct 105 ANRSSTNTEKDSRSIA------HSTSRSRSTSPANRH------RRK----EKERTRSNHR 148
Query 174 HGRVKR 179
HG +R
Sbjct 149 HGSHRR 154
> Hs13236559
Length=263
Score = 31.2 bits (69), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 26/78 (33%), Positives = 40/78 (51%), Gaps = 12/78 (15%)
Query 7 EDFKWESLKTQTNADRDYYLGASAKVGICTRGGKFEK---YDWWTK-KKEADGPSGAEDE 62
+ F WE +K T+ R+ YLG S + G+++K W+ K + GPS +E
Sbjct 14 DQFNWEDVK--TDKQRENYLGNSLMAPV----GRWQKGRDLTWYAKGRAPCAGPS--REE 65
Query 63 ELRRVKRFEQQLLEEALG 80
EL V+ E++ L ALG
Sbjct 66 ELAAVREAEREALLAALG 83
> YLL018c
Length=557
Score = 28.1 bits (61), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 23/59 (38%), Positives = 34/59 (57%), Gaps = 8/59 (13%)
Query 93 ADEEPAEPPVATTAAADDRRAAEKALKKLKKEQRKLKKLRKKEHKREAKRLRLKRERHS 151
A EE AEP D + ++KALKKL+KEQ +K+ K+E + L+L+ ER +
Sbjct 10 AVEESAEP-AQVILGEDGKPLSKKALKKLQKEQ-------EKQRKKEERALQLEAEREA 60
Lambda K H
0.313 0.130 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2878611680
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40