bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0118_orf1
Length=229
Score E
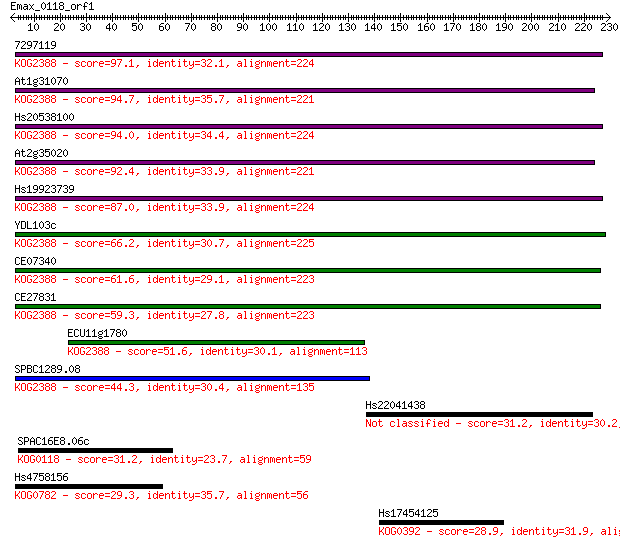
Sequences producing significant alignments: (Bits) Value
7297119 97.1 3e-20
At1g31070 94.7 2e-19
Hs20538100 94.0 2e-19
At2g35020 92.4 6e-19
Hs19923739 87.0 3e-17
YDL103c 66.2 6e-11
CE07340 61.6 1e-09
CE27831 59.3 6e-09
ECU11g1780 51.6 1e-06
SPBC1289.08 44.3 2e-04
Hs22041438 31.2 1.9
SPAC16E8.06c 31.2 2.1
Hs4758156 29.3 6.3
Hs17454125 28.9 9.6
> 7297119
Length=520
Score = 97.1 bits (240), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 72/231 (31%), Positives = 106/231 (45%), Gaps = 66/231 (28%)
Query 3 VAEYSELPPSLAE----DGAL-YAWGNVCMHYFSLAFIQAVTSNLAVYEK--RFHAAKKK 55
V EYSE+ AE DG L ++ GN+C H+FS F+Q + S YE+ + H AKKK
Sbjct 338 VVEYSEISAKTAEMRNSDGRLTFSAGNICNHFFSSNFLQKIGS---TYEQELKLHVAKKK 394
Query 56 IPELVCLDHNSGSNQRAAAETPATQDVGTPMTENPRSWALRTPTEANGWKLELFIFDAFA 115
IP + N+G TP + NG K+E F+FD F
Sbjct 395 IPFV----DNAGKRL--------------------------TPDKPNGIKIEKFVFDVFE 424
Query 116 FADRVLCLEVEREEEFAPVKVSGNIEMQALEGQGLEVASVASDTPEHAQLLLSRLHAKWV 175
FA + + +EV R+EEF+ +K S + D P A+ L RLH K++
Sbjct 425 FAQKFVAMEVPRDEEFSALKNSD---------------AAGKDCPSTARSDLHRLHKKYI 469
Query 176 AAAVSLSSLPGRLPEAPHHIFCEVSPLLSYEGEGISSGCLKGKDLSKPLLL 226
A + H CE+SP ++Y GE ++S ++GK + P+ L
Sbjct 470 EGAGGIV----------HGEVCEISPFVTYAGENLAS-HVEGKSFTSPVYL 509
> At1g31070
Length=500
Score = 94.7 bits (234), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 79/227 (34%), Positives = 99/227 (43%), Gaps = 65/227 (28%)
Query 3 VAEYSELPPSLA-----EDGAL-YAWGNVCMHYFSLAFIQAVTSNLAVYEKRFHAAKKKI 56
V EYSEL S+A G L Y W NVC+H F+L F+ V + L + +H A+KKI
Sbjct 328 VVEYSELDQSMASAINQRTGRLQYCWSNVCLHMFTLDFLNQVATGLE-KDSVYHLAEKKI 386
Query 57 PELVCLDHNSGSNQRAAAETPATQDVGTPMTENPRSWALRTPTEANGWKLELFIFDAFAF 116
P + G M G KLE FIFD+F +
Sbjct 387 PSMN----------------------GYTM----------------GLKLEQFIFDSFPY 408
Query 117 ADRVLCLEVEREEEFAPVKVSGNIEMQALEGQGLEVASVASDTPEHAQLLLSRLHAKWVA 176
A EV REEEFAPVK V DTPE A+LL+ RLH +WV
Sbjct 409 APSTALFEVLREEEFAPVK---------------NVNGSNFDTPESARLLVLRLHTRWVI 453
Query 177 AAVSLSSLPGRLPEAPHHIFCEVSPLLSYEGEGISSGCLKGKDLSKP 223
AA L +P + EVSPL SY GE + + C +G+ P
Sbjct 454 AAGGF--LTHSVPLYATGV--EVSPLCSYAGENLEAIC-RGRTFHAP 495
> Hs20538100
Length=266
Score = 94.0 bits (232), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 77/238 (32%), Positives = 103/238 (43%), Gaps = 61/238 (25%)
Query 3 VAEYSELPPSLAE----DGAL-YAWGNVCMHYFSLAFIQAVTSNLAVYEKRFHAAKKKIP 57
V EYSE+ P A+ DG+L Y GN+C H+F+ F++AVT K H A KK+P
Sbjct 63 VVEYSEISPETAQLRASDGSLLYNAGNICNHFFTRGFLKAVTREFEPLLKP-HVAVKKVP 121
Query 58 ELVCLDHNSGSNQRAAAETPATQDVGTPMTENPRSWALRTPTEANGWKLELFIFDAFAFA 117
+D L P + NG K+E F+FD F FA
Sbjct 122 ---YVDEEGN---------------------------LVKPLKPNGIKMEKFVFDVFRFA 151
Query 118 DRVLCLEVEREEEFAPVKVSGNIEMQALEGQGLEVASVASDTPEHAQLLLSRLHAKWVAA 177
LEV REEEF+P+K N E D+P A+ L H +W
Sbjct 152 KNFAALEVLREEEFSPLK---NAE------------PADRDSPRTARQALLTQHYRWALR 196
Query 178 AVS---------LSSLPGRLPEAPHHIFCEVSPLLSYEGEGISSGCLKGKDLSKPLLL 226
A + L LP P CE+SPL+SY GEG+ L+G++ PL+L
Sbjct 197 AGARFLDAHGAWLPELPSLPPNGDPPAICEISPLVSYSGEGLEV-YLQGREFQSPLIL 253
> At2g35020
Length=502
Score = 92.4 bits (228), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 75/227 (33%), Positives = 98/227 (43%), Gaps = 65/227 (28%)
Query 3 VAEYSELPPSLA-----EDGAL-YAWGNVCMHYFSLAFIQAVTSNLAVYEKRFHAAKKKI 56
V EY+EL S+A + G L Y W NVC+H F+L F+ V + L + +H A+KKI
Sbjct 330 VVEYTELDQSMASATNQQTGRLQYCWSNVCLHMFTLDFLNQVANGLE-KDSVYHLAEKKI 388
Query 57 PELVCLDHNSGSNQRAAAETPATQDVGTPMTENPRSWALRTPTEANGWKLELFIFDAFAF 116
P + + G KLE FIFD F +
Sbjct 389 PSI--------------------------------------NGDIVGLKLEQFIFDCFPY 410
Query 117 ADRVLCLEVEREEEFAPVKVSGNIEMQALEGQGLEVASVASDTPEHAQLLLSRLHAKWVA 176
A EV REEEFAPVK + DTPE A+LL+ RLH +WV
Sbjct 411 APSTALFEVLREEEFAPVKNANGSNY---------------DTPESARLLVLRLHTRWVI 455
Query 177 AAVSLSSLPGRLPEAPHHIFCEVSPLLSYEGEGISSGCLKGKDLSKP 223
AA L +P + EVSPL SY GE + + C +G+ P
Sbjct 456 AAGGF--LTHSVPLYATGV--EVSPLCSYAGENLEAIC-RGRTFHAP 497
> Hs19923739
Length=505
Score = 87.0 bits (214), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 76/240 (31%), Positives = 108/240 (45%), Gaps = 64/240 (26%)
Query 3 VAEYSELPPSLAE----DGAL-YAWGNVCMHYFSLAFIQAVTSNLAVYEKRF--HAAKKK 55
V EYSE+ + A+ DG L + GN+ H+F++ F++ V + VYE + H A+KK
Sbjct 301 VVEYSEISLATAQKRSSDGRLLFNAGNIANHFFTVPFLRDVVN---VYEPQLQHHVAQKK 357
Query 56 IPELVCLDHNSGSNQRAAAETPATQDVGTPMTENPRSWALRTPTEANGWKLELFIFDAFA 115
IP + TQ L P + NG K+E F+FD F
Sbjct 358 IPYV------------------DTQG------------QLIKPDKPNGIKMEKFVFDIFQ 387
Query 116 FADRVLCLEVEREEEFAPVKVSGNIEMQALEGQGLEVASVASDTPEHAQLLLSRLHAKWV 175
FA + + EV RE+EF+P+K N + Q D P A+ L LH WV
Sbjct 388 FAKKFVVYEVLREDEFSPLK---NADSQN-----------GKDNPTTARHALMSLHHCWV 433
Query 176 --AAAVSLSSLPGRLPEAPHH-------IFCEVSPLLSYEGEGISSGCLKGKDLSKPLLL 226
A + RLP P I CE+SPL+SY GEG+ S + K+ PL++
Sbjct 434 LNAGGHFIDENGSRLPAIPRLKDANDVPIQCEISPLISYAGEGLES-YVADKEFHAPLII 492
> YDL103c
Length=477
Score = 66.2 bits (160), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 69/234 (29%), Positives = 99/234 (42%), Gaps = 67/234 (28%)
Query 3 VAEYSELPPSLAE----DGAL-YAWGNVCMHYFSLAFIQAVTSNLAVYEKR--FHAAKKK 55
V EYSE+ LAE DG L GN+ HY+ + + +L + + +H AKKK
Sbjct 300 VIEYSEISNELAEAKDKDGLLKLRAGNIVNHYY---LVDLLKRDLDQWCENMPYHIAKKK 356
Query 56 IPELVCLDHNSGSNQRAAAETPATQDVGTPMTENPRSWALRTPTEANGWKLELFIFDAFA 115
IP D +G + PTE NG KLE FIFD F
Sbjct 357 IP---AYDSVTGKYTK--------------------------PTEPNGIKLEQFIFDVFD 387
Query 116 FA--DRVLCLEVEREEEFAPVKVSGNIEMQALEGQGLEVASVASDTPEHAQLLLSRLHAK 173
++ CLEV+R +EF+P+K G G + +D PE ++L +L
Sbjct 388 TVPLNKFGCLEVDRCKEFSPLK----------NGPGSK-----NDNPETSRLAYLKLGTS 432
Query 174 WVAAAVSLSSLPGRLPEAPHHIFCEVSPLLSYEGEGISSGCLKGKDLSKPLLLL 227
W+ A ++ + EVS LSY GE +S KGK + ++L
Sbjct 433 WLEDAGAI---------VKDGVLVEVSSKLSYAGENLSQ--FKGKVFDRSGIVL 475
> CE07340
Length=267
Score = 61.6 bits (148), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 65/228 (28%), Positives = 95/228 (41%), Gaps = 62/228 (27%)
Query 3 VAEYSELPPSLAE----DGA-LYAWGNVCMHYFSLAFIQAVTSNLAVYEKRFHAAKKKIP 57
V EYSEL LAE DG L+ G++ H+F++ F+ V S + +H A KKI
Sbjct 85 VVEYSELGAELAEQKTPDGKYLFGAGSIANHFFTMNFMDRVCSPSSRLP--YHRAHKKI- 141
Query 58 ELVCLDHNSGSNQRAAAETPATQDVGTPMTENPRSWALRTPTEANGWKLELFIFDAFAFA 117
S N++ P ENP NG KLE FIFD F +
Sbjct 142 --------SYVNEQGTIVKP----------ENP-----------NGIKLEQFIFDVFELS 172
Query 118 DRVLCLEVEREEEFAPVKVSGNIEMQALEGQGLEVASVASDTPEHAQLLLSRLHAKWVAA 177
R EV R EEF+P+K + ++ L ++ D +L L R+ AK A
Sbjct 173 KRFFIWEVARNEEFSPLKNAQSVGTDCL-------STCQRDLSNVNKLWLERVQAKVTAT 225
Query 178 AVSLSSLPGRLPEAPHHIFCEVSPLLSYEGEGISSGCLKGKDLSKPLL 225
E P ++ ++SY GE + L+ +++S L
Sbjct 226 ------------EKPIYL----KTIVSYNGESLQE--LRHREISDSAL 255
> CE27831
Length=484
Score = 59.3 bits (142), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 62/228 (27%), Positives = 94/228 (41%), Gaps = 62/228 (27%)
Query 3 VAEYSELPPSLAE----DGA-LYAWGNVCMHYFSLAFIQAVTSNLAVYEKRFHAAKKKIP 57
V EYSEL LAE DG L+ G++ H+F++ F+ V S + +H A KKI
Sbjct 302 VVEYSELGAELAEQKTPDGKYLFGAGSIANHFFTMDFMDRVCSPSSRLP--YHRAHKKIS 359
Query 58 ELVCLDHNSGSNQRAAAETPATQDVGTPMTENPRSWALRTPTEANGWKLELFIFDAFAFA 117
+ + GT + P + NG KLE FIFD F +
Sbjct 360 YV--------------------NEQGT----------IVKPEKPNGIKLEQFIFDVFELS 389
Query 118 DRVLCLEVEREEEFAPVKVSGNIEMQALEGQGLEVASVASDTPEHAQLLLSRLHAKWVAA 177
R EV R EEF+P+K + ++ L ++ D +L L R+ AK A
Sbjct 390 KRFFIWEVARNEEFSPLKNAQSVGTDCL-------STCQRDLSNVNKLWLERVQAKVTAT 442
Query 178 AVSLSSLPGRLPEAPHHIFCEVSPLLSYEGEGISSGCLKGKDLSKPLL 225
E P ++ ++SY GE + L+ +++S L
Sbjct 443 ------------EKPIYL----KTIVSYNGENLQE--LRHREISDSAL 472
> ECU11g1780
Length=335
Score = 51.6 bits (122), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 34/113 (30%), Positives = 50/113 (44%), Gaps = 35/113 (30%)
Query 23 GNVCMHYFSLAFIQAVTSNLAVYEKRFHAAKKKIPELVCLDHNSGSNQRAAAETPATQDV 82
GN+C H F +FI+ + N+ + E H A KKIP +
Sbjct 231 GNICNHIFKTSFIKKM-KNINLPE---HKAFKKIPYTI---------------------- 264
Query 83 GTPMTENPRSWALRTPTEANGWKLELFIFDAFAFADRVLCLEVEREEEFAPVK 135
S L P + NG+K E FIFD+F + + + V RE+EF+P+K
Sbjct 265 ---------SGKLIKPVKPNGFKKETFIFDSFEYTQKNGVMNVPREKEFSPLK 308
> SPBC1289.08
Length=475
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 41/145 (28%), Positives = 54/145 (37%), Gaps = 41/145 (28%)
Query 3 VAEYSELPPSLAE-----DG---ALYAWGNVCMHYFSLAFIQAVTSNLAVYEKRFHAAKK 54
V EYSE+ + DG L N+ HYFS F+Q + + + H A K
Sbjct 299 VVEYSEISDEACKATENVDGHKHLLLRAANIAYHYFSFDFLQKASLHSSTLP--IHLACK 356
Query 55 KIPELVCLDHNSGSNQRAAAETPATQDVGTPMTENPRSWALRTPTEANGWKLELFIFDAF 114
KIP H+ TP NG+KLE FIFD F
Sbjct 357 KIPFYDVTSHH-----------------------------YTTPLNPNGYKLESFIFDLF 387
Query 115 --AFADRVLCLEVEREEEFAPVKVS 137
+ C +V R F+P+K S
Sbjct 388 PSVSVENFGCFQVPRRTSFSPLKNS 412
> Hs22041438
Length=110
Score = 31.2 bits (69), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 26/89 (29%), Positives = 40/89 (44%), Gaps = 6/89 (6%)
Query 137 SGNIEM---QALEGQGLEVASVASDTPEHAQLLLSRLHAKWVAAAVSLSSLPGRLPEAPH 193
SG+IE + LE L V AS + AQ + SR +W + S LP ++ +P
Sbjct 22 SGSIEFMTQETLETWSLGVLQRASIST--AQHITSRCIKEWKCDSTSFIILPSQV-HSPK 78
Query 194 HIFCEVSPLLSYEGEGISSGCLKGKDLSK 222
H + ++ +S G L GK S+
Sbjct 79 HFYFSDDVYFAFMVYALSKGVLSGKTFSE 107
> SPAC16E8.06c
Length=438
Score = 31.2 bits (69), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 14/59 (23%), Positives = 30/59 (50%), Gaps = 5/59 (8%)
Query 4 AEYSELPPSLAEDGALYAWGNVCMHYFSLAFIQAVTSNLAVYEKRFHAAKKKIPELVCL 62
+Y +L + GA+ + + + SLAF +A+ +A +EK+FH+ + + +
Sbjct 180 GDYKDLTKHFRQFGAVDS-----IRFRSLAFSEAIPRKVAFFEKKFHSERDTVNAYIVF 233
> Hs4758156
Length=1065
Score = 29.3 bits (64), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 20/56 (35%), Positives = 25/56 (44%), Gaps = 7/56 (12%)
Query 3 VAEYSELPPSLAEDGALYAWGNVCMHYFSLAFIQAVTSNLAVYEKRFHAAKKKIPE 58
V +LPP EDG NV +YFSL F VT FH +++ PE
Sbjct 504 VERNPDLPPEELEDGVCKLPLNVFNNYFSLGFDAHVTLE-------FHESREANPE 552
> Hs17454125
Length=1849
Score = 28.9 bits (63), Expect = 9.6, Method: Compositional matrix adjust.
Identities = 15/47 (31%), Positives = 25/47 (53%), Gaps = 0/47 (0%)
Query 142 MQALEGQGLEVASVASDTPEHAQLLLSRLHAKWVAAAVSLSSLPGRL 188
+Q L+ + +V ++T + Q+L R+H A VSL LP +L
Sbjct 828 LQQLDSKRQQVQMTVTETNQEWQVLQLRVHTFAACAVVSLQQLPEKL 874
Lambda K H
0.316 0.132 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4419841864
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40