bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
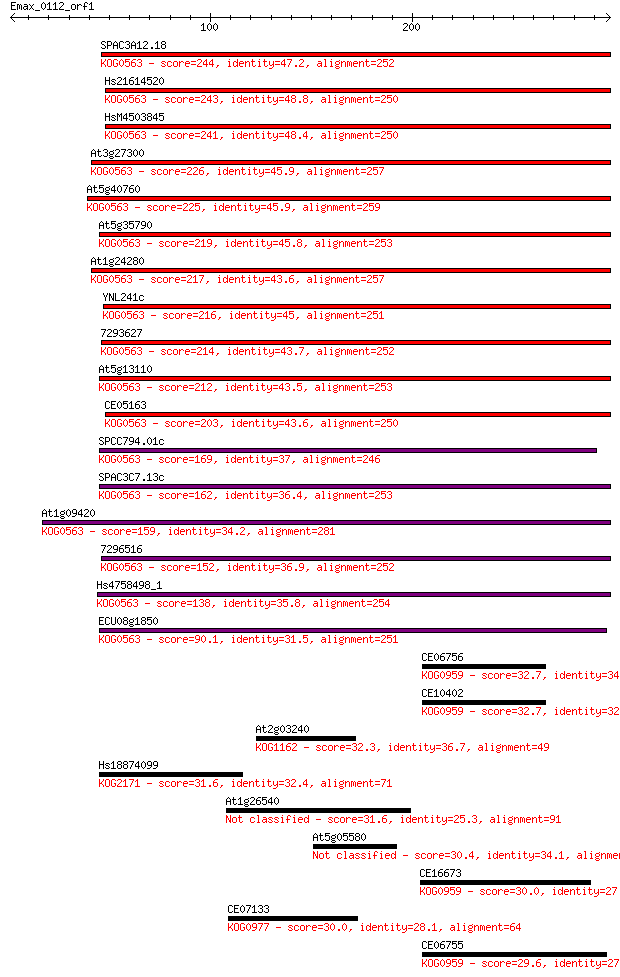
Query= Emax_0112_orf1
Length=297
Score E
Sequences producing significant alignments: (Bits) Value
SPAC3A12.18 244 2e-64
Hs21614520 243 2e-64
HsM4503845 241 1e-63
At3g27300 226 3e-59
At5g40760 225 1e-58
At5g35790 219 6e-57
At1g24280 217 3e-56
YNL241c 216 4e-56
7293627 214 2e-55
At5g13110 212 8e-55
CE05163 203 3e-52
SPCC794.01c 169 9e-42
SPAC3C7.13c 162 9e-40
At1g09420 159 6e-39
7296516 152 6e-37
Hs4758498_1 138 1e-32
ECU08g1850 90.1 5e-18
CE06756 32.7 0.86
CE10402 32.7 1.0
At2g03240 32.3 1.4
Hs18874099 31.6 1.9
At1g26540 31.6 2.1
At5g05580 30.4 4.2
CE16673 30.0 5.6
CE07133 30.0 6.2
CE06755 29.6 7.6
> SPAC3A12.18
Length=447
Score = 244 bits (622), Expect = 2e-64, Method: Compositional matrix adjust.
Identities = 119/252 (47%), Positives = 175/252 (69%), Gaps = 10/252 (3%)
Query 46 SVVVFGASGDLAKRKTYPALFSLFCEGLLPPNVHIVGFARSKLELGDFWRQIAEHLTELS 105
++VVFGASGDL+K+KT+PALFSLF EG LP ++ IVG+ARSK+E DF +I +++
Sbjct 13 AMVVFGASGDLSKKKTFPALFSLFSEGRLPKDIRIVGYARSKIEHEDFLDRITQNI---- 68
Query 106 TYFSRQVAPLSSKCEDLVQRFRSICSYICGDGYDDEVALKKLSHHLDTLEGPDGHNNRLF 165
++ S+ ++ ++ F+ CSY G YD + L+ HL EG +NR+F
Sbjct 69 -----KIDEEDSQAKEKLEEFKKRCSYYRG-SYDKPEDFEGLNSHLCEREGDRSTHNRIF 122
Query 166 YLALPPQLFALNVRSIRKHCWTQRGWNRVVVEKPFGRDSKSSEKLSNELMEVLQEKEIYR 225
YLALPP +F +++K C ++G R+V+EKPFG D KS+++L ++L + EKEIYR
Sbjct 123 YLALPPDVFVSVATNLKKKCVPEKGIARLVIEKPFGVDLKSAQELQSQLAPLFDEKEIYR 182
Query 226 IDHYLGKEMTLSIIALRFANVAFKHLFHRHNVRCVRITFKEDIGTKGRGGYFNSYGIIRD 285
IDHYLGKEM +++ LRF N HL+ ++++ V+ITFKE IGT+GRGGYF+S I+RD
Sbjct 183 IDHYLGKEMVQNLVHLRFCNPVISHLWDKNSISSVQITFKEPIGTEGRGGYFDSSTIVRD 242
Query 286 VMQNHMLQLLTL 297
++QNH++Q+LTL
Sbjct 243 IVQNHLVQILTL 254
> Hs21614520
Length=515
Score = 243 bits (621), Expect = 2e-64, Method: Compositional matrix adjust.
Identities = 122/250 (48%), Positives = 167/250 (66%), Gaps = 14/250 (5%)
Query 48 VVFGASGDLAKRKTYPALFSLFCEGLLPPNVHIVGFARSKLELGDFWRQIAEHLTELSTY 107
++ GASGDLAK+K YP ++ LF +GLLP N IVG+ARS+L + D +Q S
Sbjct 35 IIMGASGDLAKKKIYPTIWWLFRDGLLPENTFIVGYARSRLTVADIRKQ--------SEP 86
Query 108 FSRQVAPLSSKCEDLVQRFRSICSYICGDGYDDEVALKKLSHHLDTLEGPDGHNNRLFYL 167
F + K ED R SY+ G YDD + ++L+ H+D L NRLFYL
Sbjct 87 FFKATPEEKLKLEDFFAR----NSYVAGQ-YDDAASYQRLNSHMDALH-LGSQANRLFYL 140
Query 168 ALPPQLFALNVRSIRKHCWTQRGWNRVVVEKPFGRDSKSSEKLSNELMEVLQEKEIYRID 227
ALPP ++ ++I + C +Q GWNR++VEKPFGRD +SS++LSN + + +E +IYRID
Sbjct 141 ALPPTVYEAVTKNIHESCMSQIGWNRIIVEKPFGRDLQSSDRLSNHISSLFREDQIYRID 200
Query 228 HYLGKEMTLSIIALRFANVAFKHLFHRHNVRCVRITFKEDIGTKGRGGYFNSYGIIRDVM 287
HYLGKEM +++ LRFAN F +++R N+ CV +TFKE GT+GRGGYF+ +GIIRDVM
Sbjct 201 HYLGKEMVQNLMVLRFANRIFGPIWNRDNIACVILTFKEPFGTEGRGGYFDEFGIIRDVM 260
Query 288 QNHMLQLLTL 297
QNH+LQ+L L
Sbjct 261 QNHLLQMLCL 270
> HsM4503845
Length=515
Score = 241 bits (616), Expect = 1e-63, Method: Compositional matrix adjust.
Identities = 121/250 (48%), Positives = 167/250 (66%), Gaps = 14/250 (5%)
Query 48 VVFGASGDLAKRKTYPALFSLFCEGLLPPNVHIVGFARSKLELGDFWRQIAEHLTELSTY 107
++ GASGDLAK+K YP ++ LF +GLLP N IVG+ARS+L + D +Q S
Sbjct 35 IIMGASGDLAKKKIYPTIWWLFRDGLLPENTFIVGYARSRLTVADIRKQ--------SEP 86
Query 108 FSRQVAPLSSKCEDLVQRFRSICSYICGDGYDDEVALKKLSHHLDTLEGPDGHNNRLFYL 167
F + K ED R SY+ G YDD + ++L+ H++ L NRLFYL
Sbjct 87 FFKATPEEKLKLEDFFAR----NSYVAGQ-YDDAASYQRLNSHMNALH-LGSQANRLFYL 140
Query 168 ALPPQLFALNVRSIRKHCWTQRGWNRVVVEKPFGRDSKSSEKLSNELMEVLQEKEIYRID 227
ALPP ++ ++I + C +Q GWNR++VEKPFGRD +SS++LSN + + +E +IYRID
Sbjct 141 ALPPTVYEAVTKNIHESCMSQIGWNRIIVEKPFGRDLQSSDRLSNHISSLFREDQIYRID 200
Query 228 HYLGKEMTLSIIALRFANVAFKHLFHRHNVRCVRITFKEDIGTKGRGGYFNSYGIIRDVM 287
HYLGKEM +++ LRFAN F +++R N+ CV +TFKE GT+GRGGYF+ +GIIRDVM
Sbjct 201 HYLGKEMVQNLMVLRFANRIFGPIWNRDNIACVILTFKEPFGTEGRGGYFDEFGIIRDVM 260
Query 288 QNHMLQLLTL 297
QNH+LQ+L L
Sbjct 261 QNHLLQMLCL 270
> At3g27300
Length=516
Score = 226 bits (577), Expect = 3e-59, Method: Compositional matrix adjust.
Identities = 118/265 (44%), Positives = 173/265 (65%), Gaps = 18/265 (6%)
Query 41 EMHHLSVVVFGASGDLAKRKTYPALFSLFCEGLLPPN-VHIVGFARSKLELGDFWRQIAE 99
E LS++V GASGDLAK+KT+PALF+LF +G L P+ VHI G+ARSK+ + +I
Sbjct 28 ETGSLSIIVLGASGDLAKKKTFPALFNLFHQGFLNPDEVHIFGYARSKITDEELRDKIRG 87
Query 100 HLTELSTYFSRQVAPLSSKCEDLVQRFRSICSYICGDGYDDEVALKKLS----HHLDTLE 155
+L + +SK + + +F + Y+ G YD E K+L H + +
Sbjct 88 YLVDEKN---------ASKKTEALSKFLKLIKYVSGP-YDSEEGFKRLDKAILEHEISKK 137
Query 156 GPDGHNNRLFYLALPPQLFALNVRSIRKHCWTQR---GWNRVVVEKPFGRDSKSSEKLSN 212
+G + RLFYLALPP ++ + I+ C + GW R+VVEKPFG+D +S+E+LS+
Sbjct 138 TAEGSSRRLFYLALPPSVYPPVSKMIKAWCTNKSDLGGWTRIVVEKPFGKDLESAEQLSS 197
Query 213 ELMEVLQEKEIYRIDHYLGKEMTLSIIALRFANVAFKHLFHRHNVRCVRITFKEDIGTKG 272
++ + +E +IYRIDHYLGKE+ +++ LRFAN F L++R N+ V+I F+ED GT+G
Sbjct 198 QIGALFEEPQIYRIDHYLGKELVQNMLVLRFANRLFLPLWNRDNIANVQIVFREDFGTEG 257
Query 273 RGGYFNSYGIIRDVMQNHMLQLLTL 297
RGGYF+ YGIIRD++QNH+LQ+L L
Sbjct 258 RGGYFDEYGIIRDIIQNHLLQVLCL 282
> At5g40760
Length=515
Score = 225 bits (573), Expect = 1e-58, Method: Compositional matrix adjust.
Identities = 119/267 (44%), Positives = 174/267 (65%), Gaps = 19/267 (7%)
Query 39 MPEMHHLSVVVFGASGDLAKRKTYPALFSLFCEGLLPPN-VHIVGFARSKLELGDFWRQI 97
+PE LS++V GASGDLAK+KT+PALF+L+ +G L P+ VHI G+AR+K+ + +I
Sbjct 26 VPETGCLSIIVLGASGDLAKKKTFPALFNLYRQGFLNPDEVHIFGYARTKISDEELRDRI 85
Query 98 AEHLTELSTYFSRQVAPLSSKCEDLVQRFRSICSYICGDGYDDEVALKKL----SHHLDT 153
+L + + Q LS +F + Y+ G YD E ++L S H +
Sbjct 86 RGYLVDEKN--AEQAEALS--------KFLQLIKYVSGP-YDAEEGFQRLDKAISEHEIS 134
Query 154 LEGPDGHNNRLFYLALPPQLFALNVRSIRKHCWTQR---GWNRVVVEKPFGRDSKSSEKL 210
+G + RLFYLALPP ++ + I+ C + GW R+VVEKPFG+D +S+E+L
Sbjct 135 KNSTEGSSRRLFYLALPPSVYPSVCKMIKTCCMNKSDLGGWTRIVVEKPFGKDLESAEQL 194
Query 211 SNELMEVLQEKEIYRIDHYLGKEMTLSIIALRFANVAFKHLFHRHNVRCVRITFKEDIGT 270
S+++ E+ E +IYRIDHYLGKE+ +++ LRFAN F L++R N+ V+I F+ED GT
Sbjct 195 SSQIGELFDESQIYRIDHYLGKELVQNMLVLRFANRFFLPLWNRDNIENVQIVFREDFGT 254
Query 271 KGRGGYFNSYGIIRDVMQNHMLQLLTL 297
+GRGGYF+ YGIIRD++QNH+LQ+L L
Sbjct 255 EGRGGYFDEYGIIRDIIQNHLLQVLCL 281
> At5g35790
Length=576
Score = 219 bits (557), Expect = 6e-57, Method: Compositional matrix adjust.
Identities = 116/253 (45%), Positives = 164/253 (64%), Gaps = 10/253 (3%)
Query 45 LSVVVFGASGDLAKRKTYPALFSLFCEGLLPPNVHIVGFARSKLELGDFWRQIAEHLTEL 104
LS+ V GASGDLAK+K +PALF+LF EG LP + + G+AR+KL + I+ LT
Sbjct 91 LSITVVGASGDLAKKKIFPALFALFYEGCLPQDFSVFGYARTKLTHEELRDMISSTLT-- 148
Query 105 STYFSRQVAPLSSKCEDLVQRFRSICSYICGDGYDDEVALKKLSHHLDTLEGPDGHNNRL 164
R+ KC D +++F C Y G Y+ E +L+ L E +NRL
Sbjct 149 CRIDQRE------KCGDKMEQFLKRCFYHSGQ-YNSEEDFAELNKKLKEKEA-GKISNRL 200
Query 165 FYLALPPQLFALNVRSIRKHCWTQRGWNRVVVEKPFGRDSKSSEKLSNELMEVLQEKEIY 224
+YL++PP +F VR ++ GW RV+VEKPFGRDS+SS +L+ L + L E++I+
Sbjct 201 YYLSIPPNIFVDVVRCASLRASSENGWTRVIVEKPFGRDSESSGELTRCLKQYLTEEQIF 260
Query 225 RIDHYLGKEMTLSIIALRFANVAFKHLFHRHNVRCVRITFKEDIGTKGRGGYFNSYGIIR 284
RIDHYLGKE+ ++ LRF+N+ F+ L+ R+ +R V++ F ED GT+GRGGYF+ YGIIR
Sbjct 261 RIDHYLGKELVENLSVLRFSNLVFEPLWSRNYIRNVQLIFSEDFGTEGRGGYFDQYGIIR 320
Query 285 DVMQNHMLQLLTL 297
D+MQNH+LQ+L L
Sbjct 321 DIMQNHLLQILAL 333
> At1g24280
Length=599
Score = 217 bits (552), Expect = 3e-56, Method: Compositional matrix adjust.
Identities = 112/257 (43%), Positives = 162/257 (63%), Gaps = 10/257 (3%)
Query 41 EMHHLSVVVFGASGDLAKRKTYPALFSLFCEGLLPPNVHIVGFARSKLELGDFWRQIAEH 100
++ +S+ V GASGDLAK+K +PALF+L+ EG LP + I G+ARSK+ + +++
Sbjct 109 QLSTVSITVVGASGDLAKKKIFPALFALYYEGCLPEHFTIFGYARSKMTDAELRVMVSKT 168
Query 101 LTELSTYFSRQVAPLSSKCEDLVQRFRSICSYICGDGYDDEVALKKLSHHLDTLEGPDGH 160
LT ++ + C + ++ F C Y G YD + L L EG
Sbjct 169 LT---CRIDKR-----ANCGEKMEEFLKRCFYHSGQ-YDSQEHFVALDEKLKEHEG-GRL 218
Query 161 NNRLFYLALPPQLFALNVRSIRKHCWTQRGWNRVVVEKPFGRDSKSSEKLSNELMEVLQE 220
+NRLFYL++PP +F V+ + GW RV+VEKPFGRDSK+S L+ L + L+E
Sbjct 219 SNRLFYLSIPPNIFVDAVKCASSSASSVNGWTRVIVEKPFGRDSKTSAALTKSLKQYLEE 278
Query 221 KEIYRIDHYLGKEMTLSIIALRFANVAFKHLFHRHNVRCVRITFKEDIGTKGRGGYFNSY 280
+I+RIDHYLGKE+ ++ LRF+N+ F+ L+ R +R V+ F ED GT+GRGGYF++Y
Sbjct 279 DQIFRIDHYLGKELVENLSVLRFSNLIFEPLWSRQYIRNVQFIFSEDFGTEGRGGYFDNY 338
Query 281 GIIRDVMQNHMLQLLTL 297
GIIRD+MQNH+LQ+L L
Sbjct 339 GIIRDIMQNHLLQILAL 355
> YNL241c
Length=505
Score = 216 bits (551), Expect = 4e-56, Method: Compositional matrix adjust.
Identities = 113/254 (44%), Positives = 160/254 (62%), Gaps = 14/254 (5%)
Query 47 VVVFGASGDLAKRKTYPALFSLFCEGLLPPNVHIVGFARSKLELG-DFWRQIAEHLTELS 105
+ VFGASGDLAK+KT+PALF LF EG L P+ I G+ARSKL + D ++ HL +
Sbjct 14 ISVFGASGDLAKKKTFPALFGLFREGYLDPSTKIFGYARSKLSMEEDLKSRVLPHLKK-- 71
Query 106 TYFSRQVAPLSSKCEDLVQRFRSICSYICGDGYDDEVALKKLSHHLDTLEGPDGHN--NR 163
P + V++F + SYI G+ YD + +L ++ E + +R
Sbjct 72 --------PHGEADDSKVEQFFKMVSYISGN-YDTDEGFDELRTQIEKFEKSANVDVPHR 122
Query 164 LFYLALPPQLFALNVRSIRKHCWTQRGWNRVVVEKPFGRDSKSSEKLSNELMEVLQEKEI 223
LFYLALPP +F + I+ + + G RV+VEKPFG D S+ +L L + +E+E+
Sbjct 123 LFYLALPPSVFLTVAKQIKSRVYAENGITRVIVEKPFGHDLASARELQKNLGPLFKEEEL 182
Query 224 YRIDHYLGKEMTLSIIALRFANVAFKHLFHRHNVRCVRITFKEDIGTKGRGGYFNSYGII 283
YRIDHYLGKE+ +++ LRF N ++R N++ V+I+FKE GT+GRGGYF+S GII
Sbjct 183 YRIDHYLGKELVKNLLVLRFGNQFLNASWNRDNIQSVQISFKERFGTEGRGGYFDSIGII 242
Query 284 RDVMQNHMLQLLTL 297
RDVMQNH+LQ++TL
Sbjct 243 RDVMQNHLLQIMTL 256
> 7293627
Length=524
Score = 214 bits (544), Expect = 2e-55, Method: Compositional matrix adjust.
Identities = 110/252 (43%), Positives = 156/252 (61%), Gaps = 14/252 (5%)
Query 46 SVVVFGASGDLAKRKTYPALFSLFCEGLLPPNVHIVGFARSKLELGDFWRQIAEHLTELS 105
+ V+FGASGDLAK+K YP L+ L+ + LLP G+ARS L + Q ++
Sbjct 37 TFVIFGASGDLAKKKIYPTLWWLYRDDLLPKPTKFCGYARSMLTVDSIKEQCLPYM---- 92
Query 106 TYFSRQVAPLSSKCEDLVQRFRSICSYICGDGYDDEVALKKLSHHLDTLEGPDGHNNRLF 165
+V P K + F ++ Y+ G YD + L+ L+ +E + NR+F
Sbjct 93 -----KVQPHEQK---KYEEFWALNEYVSGR-YDGRTGFELLNQQLEIMENKN-KANRIF 142
Query 166 YLALPPQLFALNVRSIRKHCWTQRGWNRVVVEKPFGRDSKSSEKLSNELMEVLQEKEIYR 225
YLALPP +F +I++ C + GWNRV++EKPFGRD SS+ LS+ L + QE ++YR
Sbjct 143 YLALPPSVFEEVTVNIKQICMSVCGWNRVIIEKPFGRDDASSQALSDHLAGLFQEDQLYR 202
Query 226 IDHYLGKEMTLSIIALRFANVAFKHLFHRHNVRCVRITFKEDIGTKGRGGYFNSYGIIRD 285
IDHYLGKEM +++ +RF N ++R N+ V ITFKE GT+GRGGYF+ +GIIRD
Sbjct 203 IDHYLGKEMVQNLMTIRFGNKILSSTWNRENIASVLITFKEPFGTQGRGGYFDEFGIIRD 262
Query 286 VMQNHMLQLLTL 297
VMQNH+LQ+L+L
Sbjct 263 VMQNHLLQILSL 274
> At5g13110
Length=593
Score = 212 bits (539), Expect = 8e-55, Method: Compositional matrix adjust.
Identities = 110/254 (43%), Positives = 161/254 (63%), Gaps = 12/254 (4%)
Query 45 LSVVVFGASGDLAKRKTYPALFSLFCEGLLPPNVHIVGFARSKLELGDFWRQIAEHLTEL 104
+S+ V GASGDLAK+K +PALF+L+ EG LP + I G++RSK+ + +++ LT
Sbjct 107 VSITVVGASGDLAKKKIFPALFALYYEGCLPEHFTIFGYSRSKMTDVELRNMVSKTLT-- 164
Query 105 STYFSRQVAPLSSKCEDLVQRFRSICSYICGDGYDDEVALKKLSHHLDTLEGPDGH-NNR 163
++ + C + ++ F C Y G YD + +L L E G +NR
Sbjct 165 -CRIDKR-----ANCGEKMEEFLKRCFYHSGQ-YDSQEHFTELDKKLKEHEA--GRISNR 215
Query 164 LFYLALPPQLFALNVRSIRKHCWTQRGWNRVVVEKPFGRDSKSSEKLSNELMEVLQEKEI 223
LFYL++PP +F V+ + GW RV+VEKPFGRDS++S L+ L + L+E +I
Sbjct 216 LFYLSIPPNIFVDAVKCASTSASSVNGWTRVIVEKPFGRDSETSAALTKSLKQYLEEDQI 275
Query 224 YRIDHYLGKEMTLSIIALRFANVAFKHLFHRHNVRCVRITFKEDIGTKGRGGYFNSYGII 283
+RIDHYLGKE+ ++ LRF+N+ F+ L+ R +R V+ F ED GT+GRGGYF++YGII
Sbjct 276 FRIDHYLGKELVENLSVLRFSNLIFEPLWSRQYIRNVQFIFSEDFGTEGRGGYFDNYGII 335
Query 284 RDVMQNHMLQLLTL 297
RD+MQNH+LQ+L L
Sbjct 336 RDIMQNHLLQILAL 349
> CE05163
Length=522
Score = 203 bits (517), Expect = 3e-52, Method: Compositional matrix adjust.
Identities = 109/255 (42%), Positives = 158/255 (61%), Gaps = 20/255 (7%)
Query 48 VVFGASGDLAKRKTYPALFSLFCEGLLPPNVHIVGFARSKLELGDFWRQIAEHLTELSTY 107
V+FGASGDLAK+K YP L+ LF + LLP N+ +G+ARS L + R+ E ++
Sbjct 37 VIFGASGDLAKKKIYPTLWWLFRDNLLPVNIKFIGYARSDLTVFKL-RESFEKNCKVRE- 94
Query 108 FSRQVAPLSSKC--EDLVQRFRSICSYICGDGYDDEVALKKLSHHLDTLEGPDGHN--NR 163
+ KC +D +++ CSY+ G YD ++L +D + + NR
Sbjct 95 --------NEKCAFDDFIKK----CSYVQGQ-YDTSEGFQRLQSSIDDFQKESNNQAVNR 141
Query 164 LFYLALPPQLFALNVRSIRKHCWTQ-RGWNRVVVEKPFGRDSKSSEKLSNELMEVLQEKE 222
L+YLALPP +F + ++K+C W RV++EKPFG D KSS +LS L ++ +E +
Sbjct 142 LYYLALPPSVFNVVSTELKKNCMDHGDSWTRVIIEKPFGHDLKSSCELSTHLAKLFKEDQ 201
Query 223 IYRIDHYLGKEMTLSIIALRFANVAFKHLFHRHNVRCVRITFKEDIGTKGRGGYFNSYGI 282
IYRIDHYLGKEM +++ +RF N ++R ++ V I+FKED GT GR GYF++ GI
Sbjct 202 IYRIDHYLGKEMVQNLMVMRFGNRILAPSWNRDHIASVMISFKEDFGTGGRAGYFDTAGI 261
Query 283 IRDVMQNHMLQLLTL 297
IRDVMQNH++Q+LTL
Sbjct 262 IRDVMQNHLMQILTL 276
> SPCC794.01c
Length=475
Score = 169 bits (427), Expect = 9e-42, Method: Compositional matrix adjust.
Identities = 91/247 (36%), Positives = 143/247 (57%), Gaps = 11/247 (4%)
Query 45 LSVVVFGASGDLAKRKTYPALFSLFCEGLLPPNVHIVGFARSKLELGDFWRQIAEHLTEL 104
LS++VFGASGDLA + T+PALF+L+ ++P + I+G+ARSKL + + H+
Sbjct 2 LSIIVFGASGDLATKMTFPALFALYVRKIIPEDFQIIGYARSKLSQEAANKIVTAHIPID 61
Query 105 STYFSRQVAPLSSKCEDLVQRFRSICSYICGDGYDDEVALKKLSHHLDTLE-GPDGHNNR 163
T + Q A V+ ++ Y+ G YD + + L+ + E P R
Sbjct 62 DTVGASQKA-----LNTFVEHYK----YVPG-TYDKPESFEMLNSIIAEKETAPASECTR 111
Query 164 LFYLALPPQLFALNVRSIRKHCWTQRGWNRVVVEKPFGRDSKSSEKLSNELMEVLQEKEI 223
+FYL LPP LFA I+ R++VEKP G D KS++ + ++L + K+
Sbjct 112 IFYLVLPPHLFAPVSELIKSKAHPNGMVTRLIVEKPIGFDYKSADAILSDLSKHWSAKDT 171
Query 224 YRIDHYLGKEMTLSIIALRFANVAFKHLFHRHNVRCVRITFKEDIGTKGRGGYFNSYGII 283
+++DH+LG++M A+RFAN F+ +++R ++ VR+ F+ED G +GRGGYF GI+
Sbjct 172 FKVDHFLGEDMIDGFTAIRFANSMFEPIWNREHIESVRVDFREDFGCEGRGGYFEGAGIL 231
Query 284 RDVMQNH 290
RDV+QNH
Sbjct 232 RDVVQNH 238
> SPAC3C7.13c
Length=473
Score = 162 bits (409), Expect = 9e-40, Method: Compositional matrix adjust.
Identities = 92/257 (35%), Positives = 153/257 (59%), Gaps = 18/257 (7%)
Query 45 LSVVVFGASGDLAKRKTYPALFSLFCEGLLP-PNVHIVGFARSKLELGDFWRQIAEHLTE 103
++ +VFGASG+LA +KT+PALF LF L+ + +++G+ARSK+ +G+F I E
Sbjct 2 VTFMVFGASGNLANKKTFPALFHLFKRNLVDRSSFYVLGYARSKIPIGEFRESIRE---- 57
Query 104 LSTYFSRQVAPLSSKCEDLVQRFRSICSYICGDGYDDEVALKKLSHHLDTLEGPDGHNN- 162
V P ++ + + Q F SY G YD + + HL+++E +
Sbjct 58 -------SVKP-DTESKQVFQDFIDRVSYFSGQ-YDQSSSYVEFRKHLESVEKKADSSKA 108
Query 163 -RLFYLALPPQLFALNVRSIRKHCWTQRGWNRVVVEKPFGRDSKSSEKLSNELMEVLQEK 221
R+FY+ALPP ++ I ++ + G +R+V+EKPFG++ +S+ KL E+ + +E+
Sbjct 109 LRIFYIALPPSVYVTVSSHIYENLYLP-GKSRLVIEKPFGKNYQSAVKLKEEVHKHWKEE 167
Query 222 EIYRIDHYLGKEMTLSIIALRFAN-VAFKHLFHRHNVRCVRITFKEDIGTKGRGGYFNSY 280
EIYRIDHY K+M + LRFAN + + +RH+++ V I E G +GR GY+++
Sbjct 168 EIYRIDHYTAKDMVNNFFTLRFANSSSIDAVLNRHSIQSVEIHMYETGGCEGRIGYYDAN 227
Query 281 GIIRDVMQNHMLQLLTL 297
G++RDV+QNH+ Q+ +
Sbjct 228 GVVRDVVQNHLTQIFCI 244
> At1g09420
Length=632
Score = 159 bits (403), Expect = 6e-39, Method: Compositional matrix adjust.
Identities = 96/286 (33%), Positives = 150/286 (52%), Gaps = 32/286 (11%)
Query 17 DDFSRRRRNASRLQLRKQSTLSMPEMHHLSVVVFGASGDLAKRKTYPALFSLFCEGLLPP 76
DD S RR AS L + V GA+G+LA+ K +PALF+L+ G LP
Sbjct 143 DDLSDVRRRAS-----------------LCIAVVGATGELARGKIFPALFALYYSGYLPE 185
Query 77 NVHIVGFARSKLELGDFWRQIAEHLTELSTYFSRQVAPLSSKCEDLVQRFRSICSYICGD 136
+V I G +R L D IA LT + C + F+S YI G
Sbjct 186 DVAIFGVSRKNLTDEDLRSIIASTLTCRVDH--------QENCGGKMDAFQSRTYYING- 236
Query 137 GYDDEVALKKLSHHLDTLEGPDGHNNRLFYLALPPQLFALNVRSIRKHCWTQRGWNRVVV 196
GY++ + +L+ + +EG + NR+FYL++P + +I + RGW R++V
Sbjct 237 GYNNRDGMSRLAERMKQIEG-ESEANRIFYLSVPQEALVDVACTIGDNAQAPRGWTRIIV 295
Query 197 EKPFGRDSKSSEKLSNELMEVLQEKEIYRIDHYLGKEMTLSIIALRFANVAFKHLFHRHN 256
EKPFG +S SS +L+ L+ +EK+IYRIDH LG+ + ++ LRF+N+ F+ L++R
Sbjct 296 EKPFGFNSHSSHQLTKSLLSKFEEKQIYRIDHMLGRNLIENLTVLRFSNLVFEPLWNRTY 355
Query 257 VRCVRITFKEDIGTKGRGGYF-----NSYGIIRDVMQNHMLQLLTL 297
+R +++ F D+ + YGIIRD++ +H+LQ + L
Sbjct 356 IRNIQVVFPSDLMLFSNFLLLFLRFSDGYGIIRDIVHSHILQTIAL 401
> 7296516
Length=581
Score = 152 bits (385), Expect = 6e-37, Method: Compositional matrix adjust.
Identities = 93/259 (35%), Positives = 139/259 (53%), Gaps = 23/259 (8%)
Query 46 SVVVFGASGDLAKRKTYPALFSLFCEGLLPPNVHIVGFARSKLELGDFWRQIAEHLTELS 105
S+VVFGASG LAK+K +PAL++LF E LP I F RS L+ + QI ++ EL
Sbjct 13 SIVVFGASGGLAKKKVFPALWALFRENRLPQGTKIFTFTRSPLQTKTYRLQILPYM-ELD 71
Query 106 TYFSRQVAPLSSKCEDLVQRFRSICSYICG--DGYDDEVALKKLSHHLDTLEGPDGHN-- 161
+ + L F + + G D ++ VAL + H +T HN
Sbjct 72 KHRDPKKYNL----------FWTTVHCVQGEYDKPENYVALTEAMVHQET-----KHNQV 116
Query 162 --NRLFYLALPPQLFALNVRSIRKHCWTQRGWNRVVVEKPFGRDSKSSEKLSNELMEVLQ 219
NR+FYLALPP +F ++ + C + GWNR++VEKPF RD S + L +
Sbjct 117 RANRIFYLALPPIVFDQVTLNVSRKCSSTTGWNRIIVEKPFARDDISYKAFQTSLCNCFR 176
Query 220 EKEIYRIDHYLGKEMTLSIIALRFANVAFKHLFHRHNVRCVRITFKEDIGTK-GRGGYFN 278
E +IY +DH L +++ + ALR++N + + +V V I+ K ++ R YFN
Sbjct 177 ESQIYLMDHLLSRQVMQNFFALRYSNHLWAETLNHRHVAAVMISIKCELPVSVNRADYFN 236
Query 279 SYGIIRDVMQNHMLQLLTL 297
+GIIRD+M NHM+Q+L +
Sbjct 237 QFGIIRDLMTNHMIQMLAM 255
> Hs4758498_1
Length=557
Score = 138 bits (348), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 91/262 (34%), Positives = 139/262 (53%), Gaps = 20/262 (7%)
Query 44 HLSVVVFGASGDLAKRKTYPALFSLFCEGLLPPNVHIVGFARSKLELGDFWRQI-AEHLT 102
H+S+++ GA+GDLAK+ + LF L+ + H F + L +++ A+ L
Sbjct 25 HVSIILLGATGDLAKKYLWQGLFQLYLDEA--GRGHSFSFHGAALTAPKQGQELMAKALE 82
Query 103 ELSTYFSRQVAPLSSKCEDLVQRFRSICSYICGDGYDDEVALKKLSHHLDTLEGPDGHNN 162
LS + +AP S C + +F + Y +D AL K +E H
Sbjct 83 SLSC--PKDMAP--SHCAEHKDQFLQLSQYRQLKTAEDYQALNK------DIEAQLQHAG 132
Query 163 -----RLFYLALPPQLFALNVRSIRKHCWTQRG-WNRVVVEKPFGRDSKSSEKLSNELME 216
R+FY ++PP + R+I C G W RVV+EKPFG D S+++L+ EL
Sbjct 133 LREAGRIFYFSVPPFAYEDIARNINSSCRPGPGAWLRVVLEKPFGHDHFSAQQLATELGT 192
Query 217 VLQEKEIYRIDHYLGKEMTLSIIALRFAN-VAFKHLFHRHNVRCVRITFKEDIGTKGRGG 275
QE+E+YR+DHYLGK+ I+ R N A L++RH+V V I KE + +GR
Sbjct 193 FFQEEEMYRVDHYLGKQAVAQILPFRDQNRKALDGLWNRHHVERVEIIMKETVDAEGRTS 252
Query 276 YFNSYGIIRDVMQNHMLQLLTL 297
++ YG+IRDV+QNH+ ++LTL
Sbjct 253 FYEEYGVIRDVLQNHLTEVLTL 274
> ECU08g1850
Length=434
Score = 90.1 bits (222), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 79/253 (31%), Positives = 121/253 (47%), Gaps = 52/253 (20%)
Query 45 LSVVVFGASGDLAKRKTYPALFSLFCEGLLPPNVHIVGFARSKLELGDFWRQIAEHLTEL 104
+ VV+FG+SGDLAKRK +PAL + EG V +VG+AR+K + + +E L E+
Sbjct 1 MKVVIFGSSGDLAKRKLFPALSRIDLEG-----VGVVGYARTKYNI-----EFSEVLQEV 50
Query 105 STYFSRQVAPLSSKCEDLVQRFRSICSYICGDGYDDEVALKKLSHHLDTLEGPDGHNNRL 164
Y +P F S +YI G YDD LK++S + +
Sbjct 51 GNY-----SP----------EFLSKVTYIPGP-YDDLSKLKEVS-----------DSETV 83
Query 165 FYLALPPQLFALNVRSIRKHCWTQRGWNRVVVEKPFGRDSKSSEKLSNELMEV--LQEKE 222
Y ++P ++ R I K + G VEKP+G +S ME+ +
Sbjct 84 LYFSVPSSVYTCLFREISKLDYKVIG-----VEKPYGDSIES-------FMEIKGFDLGK 131
Query 223 IYRIDHYLGKEMTLSIIALRFANVAFKHLFHRHNVRCVRITFKEDIGTKGRGGYFNSYGI 282
IDHYL K + +++ + A + + V+ V I KE +G +GR YF+ GI
Sbjct 132 TRFIDHYLLKPLVVAMPGIIRETGAIREVMSNRYVKSVEIVSKEVLGGEGR-HYFDKNGI 190
Query 283 IRDVMQNHMLQLL 295
IRD++ +HM +LL
Sbjct 191 IRDMVLSHMGELL 203
> CE06756
Length=845
Score = 32.7 bits (73), Expect = 0.86, Method: Compositional matrix adjust.
Identities = 21/66 (31%), Positives = 34/66 (51%), Gaps = 9/66 (13%)
Query 205 KSSEKLSNELMEVLQEKE-----IYRIDHYLGKEMTLSIIALRFANVAFKHLFHRHNVRC 259
K + +LSNELM++L+ +YR +H +E L+ ++HL H+ C
Sbjct 718 KKAIQLSNELMDILKSAAPNSRLLYRNEHNPRREFQLN----NGDEYIYRHLQKTHDAGC 773
Query 260 VRITFK 265
V +TFK
Sbjct 774 VEVTFK 779
> CE10402
Length=1067
Score = 32.7 bits (73), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 20/66 (30%), Positives = 36/66 (54%), Gaps = 9/66 (13%)
Query 205 KSSEKLSNELMEVLQE-----KEIYRIDHYLGKEMTLSIIALRFANVAFKHLFHRHNVRC 259
K + +LS ELM+VL+ + +YR +H +E+ L+ ++HL H+V C
Sbjct 777 KEAIQLSKELMDVLKSAAPNSRPLYRNEHNPRRELQLN----NGDEYVYRHLQKTHDVGC 832
Query 260 VRITFK 265
V +T++
Sbjct 833 VEVTYQ 838
> At2g03240
Length=776
Score = 32.3 bits (72), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 18/49 (36%), Positives = 24/49 (48%), Gaps = 6/49 (12%)
Query 123 VQRFRSICSYICGDGYDDEVALKKLSHHLDTLEGPDGHNNRLFYLALPP 171
VQ RSI YIC G+ D H ++T D +N LF +A+ P
Sbjct 554 VQALRSIQFYICHYGWGD------YKHRINTCTESDAYNAFLFIVAVIP 596
> Hs18874099
Length=1081
Score = 31.6 bits (70), Expect = 1.9, Method: Composition-based stats.
Identities = 23/71 (32%), Positives = 38/71 (53%), Gaps = 7/71 (9%)
Query 45 LSVVVFGASGDLAKRKTYPALFSLFCEGLLPPNVHIVGFARSKLELGDFWRQIAEHLTEL 104
L+V+ GA GD +++ P L + C+GL P+ +V A + LG F + H++
Sbjct 376 LAVLSDGA-GDHIRQRLLPPLLQIVCKGLEDPS-QVVRNA-ALFALGQFSENLQPHISS- 431
Query 105 STYFSRQVAPL 115
+SR+V PL
Sbjct 432 ---YSREVMPL 439
> At1g26540
Length=695
Score = 31.6 bits (70), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 23/92 (25%), Positives = 38/92 (41%), Gaps = 12/92 (13%)
Query 108 FSRQVAPLSSKCEDLVQRFRSICSYICGDGYDDEVALKKLSHHLDTLEGPDGHNNRLFYL 167
F R V P ++ +D+V + DG+ V +KK+ +N L Y
Sbjct 68 FIRPVPPEENQQKDVVLEEGLLVDADHKDGWWTGVVVKKMED-----------DNYLVYF 116
Query 168 ALPPQLFALNVRSIRKH-CWTQRGWNRVVVEK 198
LPP + + +R H WT W + +E+
Sbjct 117 DLPPDIIQFERKQLRTHLIWTGGTWIQPEIEE 148
> At5g05580
Length=435
Score = 30.4 bits (67), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 14/42 (33%), Positives = 20/42 (47%), Gaps = 1/42 (2%)
Query 151 LDTLEGPDGHNNRLFYLALPPQLFALNVRS-IRKHCWTQRGW 191
L TL+ P + F PP ++R+ I KHCW + W
Sbjct 67 LTTLQSPSEEDTERFDPGAPPPFNLADIRAAIPKHCWVKNPW 108
> CE16673
Length=301
Score = 30.0 bits (66), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 23/90 (25%), Positives = 51/90 (56%), Gaps = 14/90 (15%)
Query 204 SKSSEKLSNELMEVLQE-----KEIYRIDHYLGKEMTLSIIALRFANVAFKHLFHRHNVR 258
K + +LS +L+++L+ + ++R +H L +E+ L+ ++HL H+V
Sbjct 17 EKETIQLSKDLIDILKSAAPSSRPLFRNEHILRREIQLN----NGDEYIYRHLQTTHDVG 72
Query 259 CVRITFKEDIGTKGRGGYFNSY-GIIRDVM 287
CV++T++ IG + Y N+ G+I++++
Sbjct 73 CVQVTYQ--IGVQ--NTYDNAVIGLIKNLI 98
> CE07133
Length=443
Score = 30.0 bits (66), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 18/64 (28%), Positives = 30/64 (46%), Gaps = 0/64 (0%)
Query 109 SRQVAPLSSKCEDLVQRFRSICSYICGDGYDDEVALKKLSHHLDTLEGPDGHNNRLFYLA 168
S + + ++ + + R RSI + G D E ++L +D L D N RLF +A
Sbjct 296 SESYSSIQNQAREEILRIRSIVNEFRGKLSDSETINQQLIKRIDDLHFQDKENLRLFEIA 355
Query 169 LPPQ 172
L +
Sbjct 356 LNEK 359
> CE06755
Length=980
Score = 29.6 bits (65), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 25/97 (25%), Positives = 49/97 (50%), Gaps = 14/97 (14%)
Query 205 KSSEKLSNELMEVLQE-----KEIYRIDHYLGKEMTLSIIALRFANVAFKHLFHRHNVRC 259
K + +LS ELM++L+ + +YR +H +E L+ ++HL H+ C
Sbjct 691 KEAIQLSKELMDILKSAAPNSRPLYRNEHNPRREFQLN----NGDEYIYRHLQKTHDAGC 746
Query 260 VRITFKEDIGTKGRGGYFNS-YGIIRDVMQNHMLQLL 295
V +T++ IG + + Y N+ G+I +++ + L
Sbjct 747 VEVTYQ--IGVQNK--YDNAVVGLIDQLIKEPVFDTL 779
Lambda K H
0.324 0.139 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6672668814
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40