bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0077_orf1
Length=189
Score E
Sequences producing significant alignments: (Bits) Value
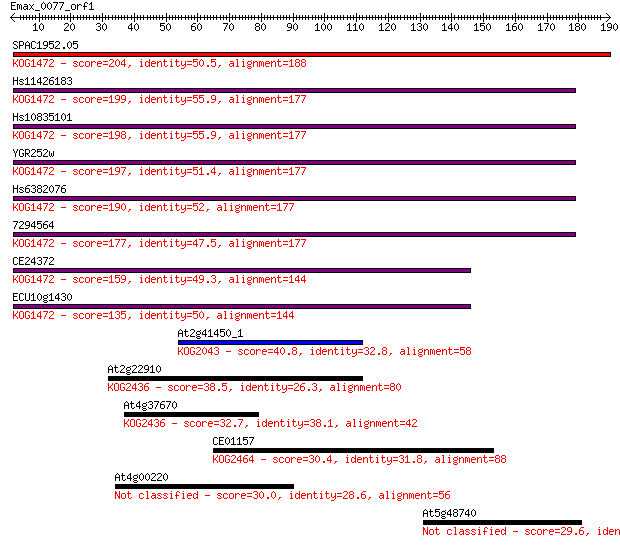
SPAC1952.05 204 6e-53
Hs11426183 199 4e-51
Hs10835101 198 4e-51
YGR252w 197 9e-51
Hs6382076 190 2e-48
7294564 177 9e-45
CE24372 159 2e-39
ECU10g1430 135 6e-32
At2g41450_1 40.8 0.002
At2g22910 38.5 0.009
At4g37670 32.7 0.48
CE01157 30.4 2.2
At4g00220 30.0 2.7
At5g48740 29.6 4.1
> SPAC1952.05
Length=454
Score = 204 bits (520), Expect = 6e-53, Method: Compositional matrix adjust.
Identities = 95/191 (49%), Positives = 138/191 (72%), Gaps = 4/191 (2%)
Query 2 LVAAKNIFSRQLPKMPREYIVRLVFDRNHYTFCLIKQQ-QIIGGCCFRPYFKQKFAEVAF 60
L KNIF +QLPKMP+EYI RL++DRNH + ++K ++GG +RP+ ++ FAE+ F
Sbjct 135 LTGLKNIFMKQLPKMPKEYITRLIYDRNHLSMTIVKDNLHVVGGITYRPFEQRGFAEIVF 194
Query 61 LAVTSAEQVKGYGTRLMNHLKEHVK-KSGIEYFLTYADNFAVGYFRKQGFTQKISMPKER 119
A+ S EQV+GYG+ LMNHLK++V+ + I++FLTYADN+A+GYF+KQGFT++I++ K
Sbjct 195 CAIASNEQVRGYGSHLMNHLKDYVRGTTTIQHFLTYADNYAIGYFKKQGFTKEITLDKSI 254
Query 120 WFGFIKDYEGGTLMECRISPKVDYLRLFAILNDQQEAVQRAILRL-KPPRVFPGLTCWQE 178
W G+IKDYEGGTLM+C + PK+ YL IL Q+ AV I R+ + V+PGL +++
Sbjct 255 WVGYIKDYEGGTLMQCTMIPKIKYLEANLILAIQKAAVVSKINRITRSNVVYPGLDVFKD 314
Query 179 DPTRVLHPSEV 189
P + PS+V
Sbjct 315 GPAHI-EPSQV 324
> Hs11426183
Length=837
Score = 199 bits (505), Expect = 4e-51, Method: Compositional matrix adjust.
Identities = 99/179 (55%), Positives = 131/179 (73%), Gaps = 2/179 (1%)
Query 2 LVAAKNIFSRQLPKMPREYIVRLVFDRNHYTFCLIKQQQIIGGCCFRPYFKQKFAEVAFL 61
LV +N+FS QLP+MP+EYI RLVFD H T LIK ++IGG CFR + Q F E+ F
Sbjct 520 LVGLQNVFSHQLPRMPKEYIARLVFDPKHKTLALIKDGRVIGGICFRMFPTQGFTEIVFC 579
Query 62 AVTSAEQVKGYGTRLMNHLKEHVKKSGIEYFLTYADNFAVGYFRKQGFTQKISMPKERWF 121
AVTS EQVKGYGT LMNHLKE+ K I YFLTYAD +A+GYF+KQGF++ I +PK R+
Sbjct 580 AVTSNEQVKGYGTHLMNHLKEYHIKHNILYFLTYADEYAIGYFKKQGFSKDIKVPKSRYL 639
Query 122 GFIKDYEGGTLMECRISPKVDYLRLFAILNDQQEAVQRAILRLKPP--RVFPGLTCWQE 178
G+IKDYEG TLMEC ++P++ Y L I+ Q+E +++ I R + +V+PGL+C++E
Sbjct 640 GYIKDYEGATLMECELNPRIPYTELSHIIKKQKEIIKKLIERKQAQIRKVYPGLSCFKE 698
> Hs10835101
Length=837
Score = 198 bits (504), Expect = 4e-51, Method: Compositional matrix adjust.
Identities = 99/179 (55%), Positives = 131/179 (73%), Gaps = 2/179 (1%)
Query 2 LVAAKNIFSRQLPKMPREYIVRLVFDRNHYTFCLIKQQQIIGGCCFRPYFKQKFAEVAFL 61
LV +N+FS QLP+MP+EYI RLVFD H T LIK ++IGG CFR + Q F E+ F
Sbjct 520 LVGLQNVFSHQLPRMPKEYIARLVFDPKHKTLALIKDGRVIGGICFRMFPTQGFTEIVFC 579
Query 62 AVTSAEQVKGYGTRLMNHLKEHVKKSGIEYFLTYADNFAVGYFRKQGFTQKISMPKERWF 121
AVTS EQVKGYGT LMNHLKE+ K I YFLTYAD +A+GYF+KQGF++ I +PK R+
Sbjct 580 AVTSNEQVKGYGTHLMNHLKEYHIKHNILYFLTYADEYAIGYFKKQGFSKDIKVPKSRYL 639
Query 122 GFIKDYEGGTLMECRISPKVDYLRLFAILNDQQEAVQRAILRLKPP--RVFPGLTCWQE 178
G+IKDYEG TLMEC ++P++ Y L I+ Q+E +++ I R + +V+PGL+C++E
Sbjct 640 GYIKDYEGATLMECELNPRIPYTELSHIIKKQKEIIKKLIERKQAQIRKVYPGLSCFKE 698
> YGR252w
Length=439
Score = 197 bits (502), Expect = 9e-51, Method: Compositional matrix adjust.
Identities = 91/180 (50%), Positives = 133/180 (73%), Gaps = 3/180 (1%)
Query 2 LVAAKNIFSRQLPKMPREYIVRLVFDRNHYTFCLIKQQ-QIIGGCCFRPYFKQKFAEVAF 60
L KNIF +QLPKMP+EYI RLV+DR+H + +I++ ++GG +RP+ K++FAE+ F
Sbjct 117 LTGLKNIFQKQLPKMPKEYIARLVYDRSHLSMAVIRKPLTVVGGITYRPFDKREFAEIVF 176
Query 61 LAVTSAEQVKGYGTRLMNHLKEHVKK-SGIEYFLTYADNFAVGYFRKQGFTQKISMPKER 119
A++S EQV+GYG LMNHLK++V+ S I+YFLTYADN+A+GYF+KQGFT++I++ K
Sbjct 177 CAISSTEQVRGYGAHLMNHLKDYVRNTSNIKYFLTYADNYAIGYFKKQGFTKEITLDKSI 236
Query 120 WFGFIKDYEGGTLMECRISPKVDYLRLFAILNDQQEAVQRAILRLKPPRVF-PGLTCWQE 178
W G+IKDYEGGTLM+C + P++ YL IL Q+ A++R I + + PGL +++
Sbjct 237 WMGYIKDYEGGTLMQCSMLPRIRYLDAGKILLLQEAALRRKIRTISKSHIVRPGLEQFKD 296
> Hs6382076
Length=832
Score = 190 bits (482), Expect = 2e-48, Method: Compositional matrix adjust.
Identities = 92/180 (51%), Positives = 130/180 (72%), Gaps = 4/180 (2%)
Query 2 LVAAKNIFSRQLPKMPREYIVRLVFDRNHYTFCLIKQQQIIGGCCFRPYFKQKFAEVAFL 61
LV +N+FS QLP+MP+EYI RLVFD H T LIK ++IGG CFR + Q F E+ F
Sbjct 515 LVGLQNVFSHQLPRMPKEYITRLVFDPKHKTLALIKDGRVIGGICFRMFPSQGFTEIVFC 574
Query 62 AVTSAEQVKGYGTRLMNHLKEHVKKSGIEYFLTYADNFAVGYFRKQGFTQKISMPKERWF 121
AVTS EQVKGYGT LMNHLKE+ K I FLTYAD +A+GYF+KQGF+++I +PK ++
Sbjct 575 AVTSNEQVKGYGTHLMNHLKEYHIKHDILNFLTYADEYAIGYFKKQGFSKEIKIPKTKYV 634
Query 122 GFIKDYEGGTLMECRISPKVDYLRLFAILNDQQEAVQRAILRLKPP---RVFPGLTCWQE 178
G+IKDYEG TLM C ++P++ Y F+++ +Q+ + + ++ K +V+PGL+C+++
Sbjct 635 GYIKDYEGATLMGCELNPRIPYTE-FSVIIKKQKEIIKKLIERKQAQIRKVYPGLSCFKD 693
> 7294564
Length=813
Score = 177 bits (450), Expect = 9e-45, Method: Compositional matrix adjust.
Identities = 84/179 (46%), Positives = 122/179 (68%), Gaps = 2/179 (1%)
Query 2 LVAAKNIFSRQLPKMPREYIVRLVFDRNHYTFCLIKQQQIIGGCCFRPYFKQKFAEVAFL 61
L+ + +F+ QLP+MPREYI +LVFD H T LIK+ Q IGG CFRP+ Q F E+ F
Sbjct 491 LLGLQLVFAYQLPEMPREYISQLVFDTKHKTLALIKENQPIGGICFRPFPSQGFTEIVFC 550
Query 62 AVTSAEQVKGYGTRLMNHLKEHVKKSGIEYFLTYADNFAVGYFRKQGFTQKISMPKERWF 121
AVT +EQVKGYGT LMNHLK++ + GI++ LT+AD A+GYF+KQGF++ I + + +
Sbjct 551 AVTMSEQVKGYGTHLMNHLKDYSIQRGIKHLLTFADCDAIGYFKKQGFSKDIKLARPVYA 610
Query 122 GFIKDYEGGTLMECRISPKVDYLRLFAILNDQQEAVQRAILRL--KPPRVFPGLTCWQE 178
G+IK+Y+ TLM C + P + + A++ Q E ++ I + + +V PGLTC++E
Sbjct 611 GYIKEYDSATLMHCELHPSIVNTQFIAVIRSQSEILKELIAQRHNEVQKVRPGLTCFKE 669
> CE24372
Length=767
Score = 159 bits (403), Expect = 2e-39, Method: Compositional matrix adjust.
Identities = 71/145 (48%), Positives = 102/145 (70%), Gaps = 1/145 (0%)
Query 2 LVAAKNIFSRQLPKMPREYIVRLVFDRNHYTFCLIKQQQ-IIGGCCFRPYFKQKFAEVAF 60
LV +N+F QLPKMP+EY+ RL+FD H ++K+ +IGG CFR + + F E+ F
Sbjct 436 LVELQNLFGAQLPKMPKEYVTRLIFDSRHQNMVILKRDMGVIGGICFRTFPSRGFVEIVF 495
Query 61 LAVTSAEQVKGYGTRLMNHLKEHVKKSGIEYFLTYADNFAVGYFRKQGFTQKISMPKERW 120
A+T+ EQVKGYGT LMNH K+++ K+ I + LTYAD FA+GYF KQGF++K+ + +
Sbjct 496 CAITAMEQVKGYGTHLMNHCKDYMIKNKIYHMLTYADEFAIGYFTKQGFSEKLEINDTVY 555
Query 121 FGFIKDYEGGTLMECRISPKVDYLR 145
G+IK+YEG TLM C + P++ Y +
Sbjct 556 QGWIKEYEGATLMGCHLHPQISYTK 580
> ECU10g1430
Length=396
Score = 135 bits (339), Expect = 6e-32, Method: Compositional matrix adjust.
Identities = 72/183 (39%), Positives = 102/183 (55%), Gaps = 41/183 (22%)
Query 2 LVAAKNIFSRQLPKMPREYIVRLVFDRNHYTFCLIKQ-QQIIGGCCFRPYFKQKFAEVAF 60
L+ K +F +QL +MP+EYI+R VFD H L+ ++I+GG C+RP+F++ F E+ F
Sbjct 44 LLGIKCLFQKQLSRMPKEYILRQVFDTKHVNMALVNSAEEIVGGICYRPFFERNFVEIVF 103
Query 61 LAVTSAEQVKGYGTRLMNHLKEHVKKSGIE------------------------------ 90
LAV QVKG G +M+ LKE VK+ +
Sbjct 104 LAVDYDFQVKGVGGFMMDLLKEVVKEEAGDCSWKSADSLLGIYEHRGRTIDDLDPLLNRT 163
Query 91 --------YFLTYADNFAVGYFRKQGFTQKISMPKERWFGFIKDYEGGTLMECRISPKVD 142
Y +TYADNFA+GYFRKQGF+ + W GFIKDYEGGT++EC +S +++
Sbjct 164 LENSPLPLYLVTYADNFAIGYFRKQGFSTDVRFGG--WIGFIKDYEGGTVVECCVSWEIN 221
Query 143 YLR 145
YL+
Sbjct 222 YLK 224
> At2g41450_1
Length=577
Score = 40.8 bits (94), Expect = 0.002, Method: Composition-based stats.
Identities = 19/58 (32%), Positives = 31/58 (53%), Gaps = 0/58 (0%)
Query 54 KFAEVAFLAVTSAEQVKGYGTRLMNHLKEHVKKSGIEYFLTYADNFAVGYFRKQGFTQ 111
++AE+ AVT Q KG+G + L + + GI +AD + G++ KQGF +
Sbjct 71 QYAEIPLAAVTYTHQKKGFGKLVYEELMKRLHSVGIRTIYCWADKESEGFWLKQGFIK 128
> At2g22910
Length=620
Score = 38.5 bits (88), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 21/81 (25%), Positives = 45/81 (55%), Gaps = 2/81 (2%)
Query 32 TFCLIKQQ-QIIGGCCFRPYFKQKFAEVAFLAVTSAEQVKGYGTRLMNHLKEHVKKSGIE 90
+F +++++ II P+F++K EVA +AV S + +G G +L++++++ G+E
Sbjct 497 SFVVVEREGHIIACAALFPFFEEKCGEVAAIAVASDCRGQGQGDKLLDYIEKKASALGLE 556
Query 91 YFLTYADNFAVGYFRKQGFTQ 111
A +F ++GF +
Sbjct 557 MLFLLTTRTA-DWFVRRGFQE 576
> At4g37670
Length=571
Score = 32.7 bits (73), Expect = 0.48, Method: Compositional matrix adjust.
Identities = 16/42 (38%), Positives = 24/42 (57%), Gaps = 0/42 (0%)
Query 37 KQQQIIGGCCFRPYFKQKFAEVAFLAVTSAEQVKGYGTRLMN 78
++ QII P+FK K EVA +AV S + +G G +L+
Sbjct 529 REGQIIACAALFPFFKDKCGEVAAIAVASDCRGQGQGDKLLG 570
> CE01157
Length=196
Score = 30.4 bits (67), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 28/94 (29%), Positives = 40/94 (42%), Gaps = 9/94 (9%)
Query 65 SAEQVKGYGTRLMNHLKEHVKKSGIEYFLTYADNFAVGYFRKQGF------TQKISMPKE 118
S EQVK +++ L KKSG + + N + RK+ T+ +
Sbjct 88 SDEQVKSVICQIIFFLVAARKKSGYSHNDFHKRNILINDTRKETICYKVEGTEYVLKTSG 147
Query 119 RWFGFIKDYEGGTLMECRISPKVDYLRLFAILND 152
W I DYEG T + + D+LRL I ND
Sbjct 148 VWVTVI-DYEGNTFDD--EGNRADFLRLDGIFND 178
> At4g00220
Length=228
Score = 30.0 bits (66), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 16/58 (27%), Positives = 29/58 (50%), Gaps = 5/58 (8%)
Query 34 CLIKQQQIIGGCCFRPYFKQKFAEVAFLAVTSAEQVKGYG--TRLMNHLKEHVKKSGI 89
C +++ + GC F PYF + F AV +V G ++L++H+ EH + +
Sbjct 21 CKFLRRKCVAGCIFAPYFDSEQGAAHFAAV---HKVFGASNVSKLLHHVPEHKRPDAV 75
> At5g48740
Length=879
Score = 29.6 bits (65), Expect = 4.1, Method: Composition-based stats.
Identities = 17/51 (33%), Positives = 25/51 (49%), Gaps = 1/51 (1%)
Query 131 TLMECRISPKVDYLRLFAILNDQQEAVQRAILRLKPPRVFPGLTC-WQEDP 180
TL + + +P+V L ++ IL EA + LK F G WQ+DP
Sbjct 320 TLRKIKFNPQVSALEVYEILQIPPEASSTTVSALKVIEQFTGQDLGWQDDP 370
Lambda K H
0.327 0.142 0.442
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3130490202
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40