bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0049_orf1
Length=122
Score E
Sequences producing significant alignments: (Bits) Value
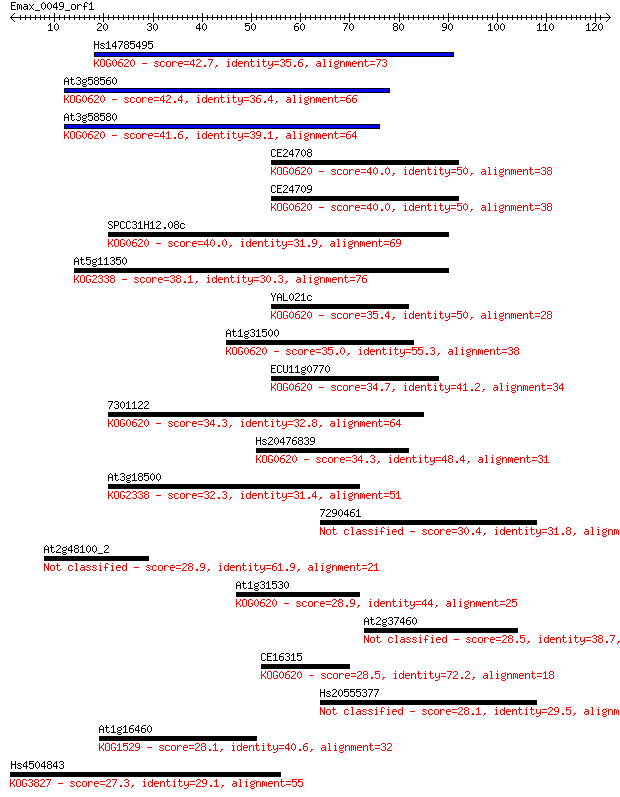
Hs14785495 42.7 1e-04
At3g58560 42.4 2e-04
At3g58580 41.6 4e-04
CE24708 40.0 0.001
CE24709 40.0 0.001
SPCC31H12.08c 40.0 0.001
At5g11350 38.1 0.004
YAL021c 35.4 0.027
At1g31500 35.0 0.030
ECU11g0770 34.7 0.040
7301122 34.3 0.052
Hs20476839 34.3 0.064
At3g18500 32.3 0.23
7290461 30.4 0.88
At2g48100_2 28.9 2.3
At1g31530 28.9 2.4
At2g37460 28.5 2.8
CE16315 28.5 3.2
Hs20555377 28.1 4.0
At1g16460 28.1 4.7
Hs4504843 27.3 6.8
> Hs14785495
Length=362
Score = 42.7 bits (99), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 26/73 (35%), Positives = 40/73 (54%), Gaps = 18/73 (24%)
Query 18 KMQHSLQLASAYALGRALLDVEENDGVEVYRQLEPIFTNYTPNFVGCLDYIFFSPFNLEL 77
++ H QL SAY EN+ L P +TNYT +F G +DYIF+S ++ +
Sbjct 235 RITHGFQLKSAY----------ENN-------LMP-YTNYTFDFKGVIDYIFYSKTHMNV 276
Query 78 VGILEPIFEQQLI 90
+G+L P+ Q L+
Sbjct 277 LGVLGPLDPQWLV 289
> At3g58560
Length=597
Score = 42.4 bits (98), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 24/70 (34%), Positives = 40/70 (57%), Gaps = 5/70 (7%)
Query 12 VLPLHWKMQHSLQLASAYA----LGRALLDVEENDGVEVYRQLEPIFTNYTPNFVGCLDY 67
+L H K+ H L L SAY+ +G ++ ++ ++ EP+FTN T +F+G LDY
Sbjct 483 ILRPHSKLTHQLPLVSAYSQFAKMGGNVITEQQRRRLDPASS-EPLFTNCTRDFIGTLDY 541
Query 68 IFFSPFNLEL 77
IF++ L +
Sbjct 542 IFYTADTLTV 551
> At3g58580
Length=605
Score = 41.6 bits (96), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 25/72 (34%), Positives = 41/72 (56%), Gaps = 12/72 (16%)
Query 12 VLPLHWKMQHSLQLASAYA--LGRALLDVEENDGVEVYRQL------EPIFTNYTPNFVG 63
+L H K+ H L L SAY+ + + ++ + G+E +R+ EP+FTN T +F+G
Sbjct 493 ILRPHTKLTHQLPLVSAYSSFVRKGIMGL----GLEQHRRRIDLNTNEPLFTNCTRDFIG 548
Query 64 CLDYIFFSPFNL 75
DYIF++ L
Sbjct 549 THDYIFYTADTL 560
> CE24708
Length=787
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 19/38 (50%), Positives = 27/38 (71%), Gaps = 1/38 (2%)
Query 54 FTNYTPNFVGCLDYIFFSPFNLELVGILEPIFEQQLIR 91
FTNYT +F G +DYIF +P +L +GIL P F+ Q ++
Sbjct 688 FTNYTLDFKGMIDYIFATPQSLARLGILGP-FDPQWVQ 724
> CE24709
Length=794
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 19/38 (50%), Positives = 27/38 (71%), Gaps = 1/38 (2%)
Query 54 FTNYTPNFVGCLDYIFFSPFNLELVGILEPIFEQQLIR 91
FTNYT +F G +DYIF +P +L +GIL P F+ Q ++
Sbjct 695 FTNYTLDFKGMIDYIFATPQSLARLGILGP-FDPQWVQ 731
> SPCC31H12.08c
Length=690
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 22/69 (31%), Positives = 34/69 (49%), Gaps = 17/69 (24%)
Query 21 HSLQLASAYALGRALLDVEENDGVEVYRQLEPIFTNYTPNFVGCLDYIFFSPFNLELVGI 80
H+ L SAY AL FTNYTP F G +D+I+++ +LE+ G+
Sbjct 598 HAFNLKSAYGESEAL-----------------SFTNYTPGFKGAIDHIWYTGNSLEVTGL 640
Query 81 LEPIFEQQL 89
L+ + + L
Sbjct 641 LKGVDKDYL 649
> At5g11350
Length=700
Score = 38.1 bits (87), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 23/76 (30%), Positives = 40/76 (52%), Gaps = 8/76 (10%)
Query 14 PLHWKMQHSLQLASAYALGRALLDVEENDGVEVYRQLEPIFTNYTPNFVGCLDYIFFSPF 73
P ++H+L+L S Y+ + + +G EP+ T+Y F+G +DYI+ S
Sbjct 600 PERTTVEHALELKSTYSEVEGQANTRDENG-------EPVVTSYHRCFMGTVDYIWRSE- 651
Query 74 NLELVGILEPIFEQQL 89
L+ V +L PI +Q +
Sbjct 652 GLQTVRVLAPIPKQAM 667
> YAL021c
Length=837
Score = 35.4 bits (80), Expect = 0.027, Method: Compositional matrix adjust.
Identities = 14/28 (50%), Positives = 20/28 (71%), Gaps = 0/28 (0%)
Query 54 FTNYTPNFVGCLDYIFFSPFNLELVGIL 81
FTN+TP+F +DYI+FS L + G+L
Sbjct 768 FTNFTPSFTDVIDYIWFSTHALRVRGLL 795
> At1g31500
Length=335
Score = 35.0 bits (79), Expect = 0.030, Method: Compositional matrix adjust.
Identities = 21/39 (53%), Positives = 25/39 (64%), Gaps = 2/39 (5%)
Query 45 EVYRQLEPIFTNYTPNFVGCLDYIFFSPFN-LELVGILE 82
EV R EP FTN TP F LDYIF SP + ++ V IL+
Sbjct 264 EVTRG-EPKFTNCTPGFTNTLDYIFISPSDFIKPVSILQ 301
> ECU11g0770
Length=493
Score = 34.7 bits (78), Expect = 0.040, Method: Composition-based stats.
Identities = 14/34 (41%), Positives = 22/34 (64%), Gaps = 1/34 (2%)
Query 54 FTNYTPNFVGCLDYIFFSPFNLELVGILEPIFEQ 87
FTN+TP F G +DYIF+ + L +L P+ ++
Sbjct 419 FTNFTPGFKGVIDYIFYGG-GISLASVLSPVEDE 451
> 7301122
Length=278
Score = 34.3 bits (77), Expect = 0.052, Method: Compositional matrix adjust.
Identities = 21/64 (32%), Positives = 33/64 (51%), Gaps = 18/64 (28%)
Query 21 HSLQLASAYALGRALLDVEENDGVEVYRQLEPIFTNYTPNFVGCLDYIFFSPFNLELVGI 80
HS +LASAY N+ + + TNYT +F G +DYIF++ + +G+
Sbjct 185 HSFKLASAY-----------NEDIMPH-------TNYTFDFKGIIDYIFYTKTGMVPLGL 226
Query 81 LEPI 84
L P+
Sbjct 227 LGPV 230
> Hs20476839
Length=139
Score = 34.3 bits (77), Expect = 0.064, Method: Compositional matrix adjust.
Identities = 15/31 (48%), Positives = 19/31 (61%), Gaps = 0/31 (0%)
Query 51 EPIFTNYTPNFVGCLDYIFFSPFNLELVGIL 81
EP +TNY F GCLDYIF LE+ ++
Sbjct 76 EPAYTNYVGGFHGCLDYIFIDLNALEVEQVI 106
> At3g18500
Length=451
Score = 32.3 bits (72), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 16/51 (31%), Positives = 29/51 (56%), Gaps = 7/51 (13%)
Query 21 HSLQLASAYALGRALLDVEENDGVEVYRQLEPIFTNYTPNFVGCLDYIFFS 71
H L+L S+YA + + ++ G EP+ T+Y F+G +DY+++S
Sbjct 363 HPLKLNSSYASVKGSANTRDSVG-------EPLATSYHSKFLGTVDYLWYS 406
> 7290461
Length=788
Score = 30.4 bits (67), Expect = 0.88, Method: Composition-based stats.
Identities = 14/44 (31%), Positives = 26/44 (59%), Gaps = 0/44 (0%)
Query 64 CLDYIFFSPFNLELVGILEPIFEQQLIREGSSSSSATAKTYCLC 107
C + F+PF++ LV ++ F+Q I + +SS A ++ Y +C
Sbjct 337 CKVFDLFTPFSVGLVYLMYKCFQQIAIIKPNSSRPANSERYLVC 380
> At2g48100_2
Length=1208
Score = 28.9 bits (63), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 13/21 (61%), Positives = 14/21 (66%), Gaps = 0/21 (0%)
Query 8 QHKEVLPLHWKMQHSLQLASA 28
Q K V PLHW +Q L LASA
Sbjct 3 QEKNVDPLHWALQLRLTLASA 23
> At1g31530
Length=283
Score = 28.9 bits (63), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 11/25 (44%), Positives = 16/25 (64%), Gaps = 0/25 (0%)
Query 47 YRQLEPIFTNYTPNFVGCLDYIFFS 71
+ + EP FTN P F LDY+F++
Sbjct 217 FTKGEPRFTNNVPGFAETLDYMFYT 241
> At2g37460
Length=380
Score = 28.5 bits (62), Expect = 2.8, Method: Composition-based stats.
Identities = 12/31 (38%), Positives = 19/31 (61%), Gaps = 0/31 (0%)
Query 73 FNLELVGILEPIFEQQLIREGSSSSSATAKT 103
F + L+G+LEP+ +Q L G ++AT T
Sbjct 79 FKISLLGLLEPVIDQNLYYLGMKYTTATFAT 109
> CE16315
Length=628
Score = 28.5 bits (62), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 13/23 (56%), Positives = 16/23 (69%), Gaps = 5/23 (21%)
Query 52 PIFTNYTPN-----FVGCLDYIF 69
P +TNYT + FVGCLDYI+
Sbjct 564 PEYTNYTASSQKDGFVGCLDYIW 586
> Hs20555377
Length=835
Score = 28.1 bits (61), Expect = 4.0, Method: Composition-based stats.
Identities = 13/44 (29%), Positives = 25/44 (56%), Gaps = 0/44 (0%)
Query 64 CLDYIFFSPFNLELVGILEPIFEQQLIREGSSSSSATAKTYCLC 107
C + F+PF++ LV +L FE+ + + +S A ++ Y +C
Sbjct 403 CKTFDLFTPFSVGLVYLLYCCFERVCLFKPITSRPANSERYVVC 446
> At1g16460
Length=318
Score = 28.1 bits (61), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 13/32 (40%), Positives = 19/32 (59%), Gaps = 0/32 (0%)
Query 19 MQHSLQLASAYALGRALLDVEENDGVEVYRQL 50
++H L A+A G + L +E NDGV VY +
Sbjct 79 LRHMLPSEEAFAAGCSALGIENNDGVVVYDGM 110
> Hs4504843
Length=423
Score = 27.3 bits (59), Expect = 6.8, Method: Composition-based stats.
Identities = 16/55 (29%), Positives = 23/55 (41%), Gaps = 0/55 (0%)
Query 1 HQSVNPQQHKEVLPLHWKMQHSLQLASAYALGRALLDVEENDGVEVYRQLEPIFT 55
HQ P+Q ++ LP H + + Y +V + E YR L IFT
Sbjct 28 HQPKLPKQARDDLPRHISRDRTKRKIQRYVRKDGKCNVHHGNVRETYRYLTDIFT 82
Lambda K H
0.321 0.134 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1194805952
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40