bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0043_orf1
Length=145
Score E
Sequences producing significant alignments: (Bits) Value
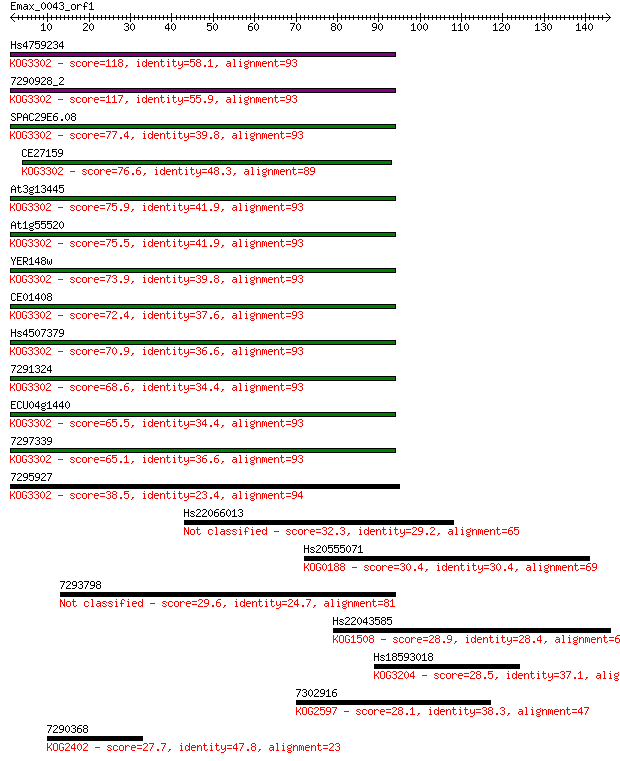
Hs4759234 118 3e-27
7290928_2 117 9e-27
SPAC29E6.08 77.4 8e-15
CE27159 76.6 1e-14
At3g13445 75.9 3e-14
At1g55520 75.5 3e-14
YER148w 73.9 1e-13
CE01408 72.4 3e-13
Hs4507379 70.9 9e-13
7291324 68.6 4e-12
ECU04g1440 65.5 3e-11
7297339 65.1 4e-11
7295927 38.5 0.004
Hs22066013 32.3 0.35
Hs20555071 30.4 1.1
7293798 29.6 2.2
Hs22043585 28.9 3.1
Hs18593018 28.5 5.1
7302916 28.1 6.7
7290368 27.7 8.3
> Hs4759234
Length=186
Score = 118 bits (296), Expect = 3e-27, Method: Compositional matrix adjust.
Identities = 54/93 (58%), Positives = 69/93 (74%), Gaps = 0/93 (0%)
Query 1 LGFRVKFCQYRVVNVLGACRLPFGIKVPEFSTEHRKLASYELELHPGATYRIKELKAVLK 60
LGF+V F ++VVNVL C +PF I++PEF+ +R ASYE ELHP YRIK L+A L+
Sbjct 91 LGFQVIFTDFKVVNVLAVCNMPFEIRLPEFTKNNRPHASYEPELHPAVCYRIKSLRATLQ 150
Query 61 IFQTGSITITAPSIQNVQLAVEQIYPLVYEFRK 93
IF TGSIT+T P+++ V AVEQIYP V+E RK
Sbjct 151 IFSTGSITVTGPNVKAVATAVEQIYPFVFESRK 183
> 7290928_2
Length=184
Score = 117 bits (292), Expect = 9e-27, Method: Compositional matrix adjust.
Identities = 52/95 (54%), Positives = 72/95 (75%), Gaps = 2/95 (2%)
Query 1 LGFRVKFCQYRVVNVLGACRLPFGIKVPEFSTEHRKLASYELELHPGATYRIK--ELKAV 58
LGF +F +R+VNVLG C +P+ IK+ FS HR+ ASYE ELHPG TY+++ + KA
Sbjct 89 LGFPTRFLNFRIVNVLGTCSMPWAIKIVNFSERHRENASYEPELHPGVTYKMRDPDPKAT 148
Query 59 LKIFQTGSITITAPSIQNVQLAVEQIYPLVYEFRK 93
LKIF TGS+T+TA S+ +V+ A++ IYPLV++FRK
Sbjct 149 LKIFSTGSVTVTAASVNHVESAIQHIYPLVFDFRK 183
> SPAC29E6.08
Length=231
Score = 77.4 bits (189), Expect = 8e-15, Method: Compositional matrix adjust.
Identities = 37/94 (39%), Positives = 57/94 (60%), Gaps = 1/94 (1%)
Query 1 LGFRVKFCQYRVVNVLGACRLPFGIKVPEFSTEHRKLASYELELHPGATYRIKELKAVLK 60
LGF KF +++ N++G+C + F I++ + H +SYE EL PG YR+ + K VL
Sbjct 137 LGFNAKFTDFKIQNIVGSCDVKFPIRLEGLAYSHGTFSSYEPELFPGLIYRMVKPKVVLL 196
Query 61 IFQTGSITITAPSI-QNVQLAVEQIYPLVYEFRK 93
IF +G I +T + + + A E IYP++ EFRK
Sbjct 197 IFVSGKIVLTGAKVREEIYQAFEAIYPVLSEFRK 230
> CE27159
Length=505
Score = 76.6 bits (187), Expect = 1e-14, Method: Composition-based stats.
Identities = 43/90 (47%), Positives = 56/90 (62%), Gaps = 1/90 (1%)
Query 4 RVKFCQYRVVNVLGACRLPFGIKVPEFSTEHRKLASYELELHPGATYRIKELKAVLKIFQ 63
RV YRV NVL CRLPFGIK+ E + ++ ++YE EL G +R KA L+I
Sbjct 352 RVSIRNYRVNNVLATCRLPFGIKIEEVAAKYPSESTYEPELSVGLVWRSVTPKATLRIHT 411
Query 64 TGSITIT-APSIQNVQLAVEQIYPLVYEFR 92
TGSIT+T A S +V + +IYP+V EFR
Sbjct 412 TGSITVTGAQSEADVLEVLSKIYPIVLEFR 441
> At3g13445
Length=200
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 39/94 (41%), Positives = 57/94 (60%), Gaps = 1/94 (1%)
Query 1 LGFRVKFCQYRVVNVLGACRLPFGIKVPEFSTEHRKLASYELELHPGATYRIKELKAVLK 60
LGF KF +++ N++G+C + F I++ + H +SYE EL PG YR+K K VL
Sbjct 104 LGFPAKFKDFKIQNIVGSCDVKFPIRLEGLAYSHAAFSSYEPELFPGLIYRMKVPKIVLL 163
Query 61 IFQTGSITITAPSIQN-VQLAVEQIYPLVYEFRK 93
IF +G I IT +++ A E IYP++ EFRK
Sbjct 164 IFVSGKIVITGAKMRDETYKAFENIYPVLSEFRK 197
> At1g55520
Length=200
Score = 75.5 bits (184), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 39/94 (41%), Positives = 56/94 (59%), Gaps = 1/94 (1%)
Query 1 LGFRVKFCQYRVVNVLGACRLPFGIKVPEFSTEHRKLASYELELHPGATYRIKELKAVLK 60
LGF KF +++ N++G+C + F I++ + H +SYE EL PG YR+K K VL
Sbjct 104 LGFPAKFKDFKIQNIVGSCDVKFPIRLEGLAYSHSAFSSYEPELFPGLIYRMKLPKIVLL 163
Query 61 IFQTGSITITAPSI-QNVQLAVEQIYPLVYEFRK 93
IF +G I IT + + A E IYP++ EFRK
Sbjct 164 IFVSGKIVITGAKMREETYTAFENIYPVLREFRK 197
> YER148w
Length=240
Score = 73.9 bits (180), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 37/94 (39%), Positives = 57/94 (60%), Gaps = 1/94 (1%)
Query 1 LGFRVKFCQYRVVNVLGACRLPFGIKVPEFSTEHRKLASYELELHPGATYRIKELKAVLK 60
+GF KF +++ N++G+C + F I++ + H +SYE EL PG YR+ + K VL
Sbjct 146 IGFAAKFTDFKIQNIVGSCDVKFPIRLEGLAFSHGTFSSYEPELFPGLIYRMVKPKIVLL 205
Query 61 IFQTGSITIT-APSIQNVQLAVEQIYPLVYEFRK 93
IF +G I +T A + + A E IYP++ EFRK
Sbjct 206 IFVSGKIVLTGAKQREEIYQAFEAIYPVLSEFRK 239
> CE01408
Length=340
Score = 72.4 bits (176), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 35/94 (37%), Positives = 59/94 (62%), Gaps = 1/94 (1%)
Query 1 LGFRVKFCQYRVVNVLGACRLPFGIKVPEFSTEHRKLASYELELHPGATYRIKELKAVLK 60
LGF+ KF ++ V N++G+C + F I++ H + ++YE EL PG YR+ + + VL
Sbjct 247 LGFQAKFTEFMVQNMVGSCDVRFPIQLEGLCITHSQFSTYEPELFPGLIYRMVKPRVVLL 306
Query 61 IFQTGSITIT-APSIQNVQLAVEQIYPLVYEFRK 93
IF +G + IT A + +++ A QIYP++ F+K
Sbjct 307 IFVSGKVVITGAKTKRDIDEAFGQIYPILKGFKK 340
> Hs4507379
Length=339
Score = 70.9 bits (172), Expect = 9e-13, Method: Compositional matrix adjust.
Identities = 34/94 (36%), Positives = 57/94 (60%), Gaps = 1/94 (1%)
Query 1 LGFRVKFCQYRVVNVLGACRLPFGIKVPEFSTEHRKLASYELELHPGATYRIKELKAVLK 60
LGF KF +++ N++G+C + F I++ H++ +SYE EL PG YR+ + + VL
Sbjct 244 LGFPAKFLDFKIQNMVGSCDVKFPIRLEGLVLTHQQFSSYEPELFPGLIYRMIKPRIVLL 303
Query 61 IFQTGSITITAPSIQ-NVQLAVEQIYPLVYEFRK 93
IF +G + +T ++ + A E IYP++ FRK
Sbjct 304 IFVSGKVVLTGAKVRAEIYEAFENIYPILKGFRK 337
> 7291324
Length=353
Score = 68.6 bits (166), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 32/94 (34%), Positives = 56/94 (59%), Gaps = 1/94 (1%)
Query 1 LGFRVKFCQYRVVNVLGACRLPFGIKVPEFSTEHRKLASYELELHPGATYRIKELKAVLK 60
LGF KF +++ N++G+C + F I++ H +SYE EL PG YR+ + VL
Sbjct 258 LGFPAKFLDFKIQNMVGSCDVKFPIRLEGLVLTHCNFSSYEPELFPGLIYRMVRPRIVLL 317
Query 61 IFQTGSITITAPSI-QNVQLAVEQIYPLVYEFRK 93
IF +G + +T + Q + A ++I+P++ +F+K
Sbjct 318 IFVSGKVVLTGAKVRQEIYDAFDKIFPILKKFKK 351
> ECU04g1440
Length=198
Score = 65.5 bits (158), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 32/94 (34%), Positives = 53/94 (56%), Gaps = 1/94 (1%)
Query 1 LGFRVKFCQYRVVNVLGACRLPFGIKVPEFSTEHRKLASYELELHPGATYRIKELKAVLK 60
LGF F +++ N++ +C + F I++ + H SYE EL PG YR+ + K VL
Sbjct 104 LGFNATFDDFKIQNIVSSCDIKFSIRLEGLAYAHSNYCSYEPELFPGLIYRMVKPKIVLL 163
Query 61 IFQTGSITITAPSIQ-NVQLAVEQIYPLVYEFRK 93
IF +G I +T ++ ++ A IYP++ + RK
Sbjct 164 IFVSGKIVLTGAKVRDDIYQAFNNIYPVLIQHRK 197
> 7297339
Length=224
Score = 65.1 bits (157), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 34/94 (36%), Positives = 55/94 (58%), Gaps = 1/94 (1%)
Query 1 LGFRVKFCQYRVVNVLGACRLPFGIKVPEFSTEHRKLASYELELHPGATYRIKELKAVLK 60
LGF VKF +Y++ N++ L F I++ + H + +SYE E+ PG YR+ + + VL
Sbjct 130 LGFPVKFMEYKLQNIVATVDLRFPIRLENLNHVHGQFSSYEPEMFPGLIYRMVKPRIVLL 189
Query 61 IFQTGSITIT-APSIQNVQLAVEQIYPLVYEFRK 93
IF G + T A S +++ +E I P++ FRK
Sbjct 190 IFVNGKVVFTGAKSRKDIMDCLEAISPILLSFRK 223
> 7295927
Length=294
Score = 38.5 bits (88), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 22/95 (23%), Positives = 45/95 (47%), Gaps = 1/95 (1%)
Query 1 LGFRVKFCQYRVVNVLGACRLPFGIKVPEFSTEHRKLASYELELHPGATYRIKELKAVLK 60
LG+ + + + V +PF + + +F E+ + Y+ +P Y++ +
Sbjct 166 LGYAPRLRRLKTNAVNATFSVPFNLNLRQFHLENPVVTRYDTSKYPFLVYKMMGTTVEIA 225
Query 61 IFQTGS-ITITAPSIQNVQLAVEQIYPLVYEFRKP 94
IF TG I + A +++ +LA+ I P +Y + P
Sbjct 226 IFPTGYVIVLFATTMEITKLAIAHILPTLYRLKDP 260
> Hs22066013
Length=138
Score = 32.3 bits (72), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 19/65 (29%), Positives = 34/65 (52%), Gaps = 4/65 (6%)
Query 43 ELHPGATYRIKELKAVLKIFQTGSITITAPSIQNVQLAVEQIYPLVYEFRKPRPQLAINS 102
LHPG ++LK +L + GSI I A S+ ++ L +E + + R+P ++I
Sbjct 78 HLHPGTNLGCRKLKCMLPLIMNGSI-IFAESLPSLSLQIESA---IQDGRRPHSTISIKR 133
Query 103 STSAA 107
+ A+
Sbjct 134 ANEAS 138
> Hs20555071
Length=985
Score = 30.4 bits (67), Expect = 1.1, Method: Composition-based stats.
Identities = 21/73 (28%), Positives = 34/73 (46%), Gaps = 4/73 (5%)
Query 72 PSIQNVQLAVEQIYPLVYEFRKPRPQLAINSSTSAAAASSAAGGGNNSPEGVGDDLT--- 128
PS+ V + Q P+ PR ++A + + AGG +N E VG DL+
Sbjct 70 PSLLFVNAGMNQFKPIFLGTVDPRSEMAGFRRVANSQKCVRAGGHHNDLEDVGRDLSHHT 129
Query 129 -IDELRNYSDGGD 140
+ L N++ GG+
Sbjct 130 FFEMLGNWAFGGE 142
> 7293798
Length=759
Score = 29.6 bits (65), Expect = 2.2, Method: Composition-based stats.
Identities = 20/88 (22%), Positives = 35/88 (39%), Gaps = 16/88 (18%)
Query 13 VNVLGACRLPFGIKVPEF-------STEHRKLASYELELHPGATYRIKELKAVLKIFQTG 65
+ ++ P+GI +PEF H K Y + PG R+ E TG
Sbjct 669 LRLISCAHFPWGISLPEFIESCAICPEPHMKYVYYVDKTIPGVVARVHE---------TG 719
Query 66 SITITAPSIQNVQLAVEQIYPLVYEFRK 93
I + A + ++++YP+ R+
Sbjct 720 MIQVFAMTTGEADNMLKKVYPITANHRR 747
> Hs22043585
Length=250
Score = 28.9 bits (63), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 19/67 (28%), Positives = 33/67 (49%), Gaps = 3/67 (4%)
Query 79 LAVEQIYPLVYEFRKPRPQLAINSSTSAAAASSAAGGGNNSPEGVGDDLTIDELRNYSDG 138
+A ++ PL + +KPRP A+ ++A+A G E + IDE++N D
Sbjct 1 MAPKRQSPLPLQKKKPRPPPALGLEETSASAGLPKKGEKEQQEAIEH---IDEVQNEIDR 57
Query 139 GDSDDSD 145
+ DS+
Sbjct 58 LNEQDSE 64
> Hs18593018
Length=105
Score = 28.5 bits (62), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 13/35 (37%), Positives = 18/35 (51%), Gaps = 0/35 (0%)
Query 89 YEFRKPRPQLAINSSTSAAAASSAAGGGNNSPEGV 123
Y FR P+P L + + AA A G + P+GV
Sbjct 71 YHFRAPQPHLLADRARYAAPQDQARPGFSGPPQGV 105
> 7302916
Length=526
Score = 28.1 bits (61), Expect = 6.7, Method: Composition-based stats.
Identities = 18/49 (36%), Positives = 25/49 (51%), Gaps = 2/49 (4%)
Query 70 TAPSIQNVQLA--VEQIYPLVYEFRKPRPQLAINSSTSAAAASSAAGGG 116
T I+NV A V+ PL+Y +P+L I+ T A + A GGG
Sbjct 361 TTMGIKNVDKAPVVQLADPLLYAQATYKPKLVIDIGTVAMGVNYAVGGG 409
> 7290368
Length=261
Score = 27.7 bits (60), Expect = 8.3, Method: Compositional matrix adjust.
Identities = 11/23 (47%), Positives = 14/23 (60%), Gaps = 0/23 (0%)
Query 10 YRVVNVLGACRLPFGIKVPEFST 32
+R+ VLG C L FG KV + T
Sbjct 98 HRIAEVLGICELDFGYKVEQIPT 120
Lambda K H
0.316 0.134 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1712413322
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40