bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0012_orf1
Length=204
Score E
Sequences producing significant alignments: (Bits) Value
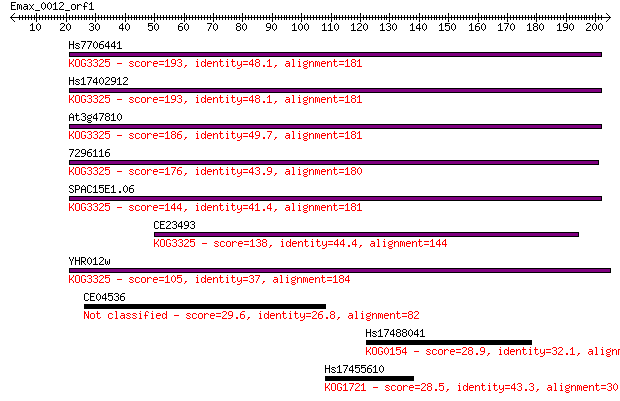
Hs7706441 193 2e-49
Hs17402912 193 2e-49
At3g47810 186 3e-47
7296116 176 3e-44
SPAC15E1.06 144 1e-34
CE23493 138 7e-33
YHR012w 105 7e-23
CE04536 29.6 4.3
Hs17488041 28.9 6.9
Hs17455610 28.5 9.1
> Hs7706441
Length=182
Score = 193 bits (491), Expect = 2e-49, Method: Compositional matrix adjust.
Identities = 87/181 (48%), Positives = 124/181 (68%), Gaps = 0/181 (0%)
Query 21 LVLIIGDFHVPQRAVDLPPCFKELLHTDKIRHVLCTGNVGSESVLEFLRGIAGSVHIVKG 80
LVL++GD H+P R LP FK+LL KI+H+LCTGN+ ++ ++L+ +AG VHIV+G
Sbjct 2 LVLVLGDLHIPHRCNSLPAKFKKLLVPGKIQHILCTGNLCTKESYDYLKTLAGDVHIVRG 61
Query 81 DMDEGMDFPVYKILQFGEFKVALLHGHQVIPWGDADALLQWQRRLDCDIVVSGHTHSNSV 140
D DE +++P K++ G+FK+ L+HGHQVIPWGD +L QR+ D DI++SGHTH
Sbjct 62 DFDENLNYPEQKVVTVGQFKIGLIHGHQVIPWGDMASLALLQRQFDVDILISGHTHKFEA 121
Query 141 REVEGRFFINPGSATGAYQPWAPSPTPSFMLMALQGASVVLYIYEERDGKAEVVMSEFSK 200
E E +F+INPGSATGAY + PSF+LM +Q ++VV Y+Y+ +V E+ K
Sbjct 122 FEHENKFYINPGSATGAYNALETNIIPSFVLMDIQASTVVTYVYQLIGDDVKVERIEYKK 181
Query 201 P 201
P
Sbjct 182 P 182
> Hs17402912
Length=186
Score = 193 bits (491), Expect = 2e-49, Method: Compositional matrix adjust.
Identities = 87/181 (48%), Positives = 124/181 (68%), Gaps = 0/181 (0%)
Query 21 LVLIIGDFHVPQRAVDLPPCFKELLHTDKIRHVLCTGNVGSESVLEFLRGIAGSVHIVKG 80
LVL++GD H+P R LP FK+LL KI+H+LCTGN+ ++ ++L+ +AG VHIV+G
Sbjct 6 LVLVLGDLHIPHRCNSLPAKFKKLLVPGKIQHILCTGNLCTKESYDYLKTLAGDVHIVRG 65
Query 81 DMDEGMDFPVYKILQFGEFKVALLHGHQVIPWGDADALLQWQRRLDCDIVVSGHTHSNSV 140
D DE +++P K++ G+FK+ L+HGHQVIPWGD +L QR+ D DI++SGHTH
Sbjct 66 DFDENLNYPEQKVVTVGQFKIGLIHGHQVIPWGDMASLALLQRQFDVDILISGHTHKFEA 125
Query 141 REVEGRFFINPGSATGAYQPWAPSPTPSFMLMALQGASVVLYIYEERDGKAEVVMSEFSK 200
E E +F+INPGSATGAY + PSF+LM +Q ++VV Y+Y+ +V E+ K
Sbjct 126 FEHENKFYINPGSATGAYNALETNIIPSFVLMDIQASTVVTYVYQLIGDDVKVERIEYKK 185
Query 201 P 201
P
Sbjct 186 P 186
> At3g47810
Length=190
Score = 186 bits (472), Expect = 3e-47, Method: Compositional matrix adjust.
Identities = 90/181 (49%), Positives = 119/181 (65%), Gaps = 0/181 (0%)
Query 21 LVLIIGDFHVPQRAVDLPPCFKELLHTDKIRHVLCTGNVGSESVLEFLRGIAGSVHIVKG 80
LVL +GD HVP RA DLPP FK +L KI+H++CTGN+ + + ++L+ I +HIV+G
Sbjct 3 LVLALGDLHVPHRAADLPPKFKSMLVPGKIQHIICTGNLCIKEIHDYLKTICPDLHIVRG 62
Query 81 DMDEGMDFPVYKILQFGEFKVALLHGHQVIPWGDADALLQWQRRLDCDIVVSGHTHSNSV 140
+ DE +P K L G+FK+ L HGHQVIPWGD D+L QR+L DI+V+GHTH +
Sbjct 63 EFDEDARYPENKTLTIGQFKLGLCHGHQVIPWGDLDSLAMLQRQLGVDILVTGHTHQFTA 122
Query 141 REVEGRFFINPGSATGAYQPWAPSPTPSFMLMALQGASVVLYIYEERDGKAEVVMSEFSK 200
+ EG INPGSATGAY PSF+LM + G V+Y+YE DG+ +V EF K
Sbjct 123 YKHEGGVVINPGSATGAYSSINQDVNPSFVLMDIDGFRAVVYVYELIDGEVKVDKIEFKK 182
Query 201 P 201
P
Sbjct 183 P 183
> 7296116
Length=182
Score = 176 bits (445), Expect = 3e-44, Method: Compositional matrix adjust.
Identities = 79/180 (43%), Positives = 119/180 (66%), Gaps = 0/180 (0%)
Query 21 LVLIIGDFHVPQRAVDLPPCFKELLHTDKIRHVLCTGNVGSESVLEFLRGIAGSVHIVKG 80
LVL++GD H+P R LP FK+LL +I H+L TGN+ ++ ++L+ +A VHIV+G
Sbjct 2 LVLVLGDLHIPHRCSSLPAKFKKLLVPGRIHHILATGNICTKESYDYLKSLANDVHIVRG 61
Query 81 DMDEGMDFPVYKILQFGEFKVALLHGHQVIPWGDADALLQWQRRLDCDIVVSGHTHSNSV 140
D DE + +P K++ G+F++ L HGHQV+P GD +AL QR+LD DI+++GHT+
Sbjct 62 DFDENLTYPEQKVVTVGQFRIGLCHGHQVVPRGDPEALALIQRQLDVDILITGHTYKFEA 121
Query 141 REVEGRFFINPGSATGAYQPWAPSPTPSFMLMALQGASVVLYIYEERDGKAEVVMSEFSK 200
E +F+INPGSATGA+ P + PSF+LM +Q +VV Y+Y+ + +V E+ K
Sbjct 122 YEHGNKFYINPGSATGAFNPLDTNVVPSFVLMDIQSTTVVTYVYQLIGDEVKVERIEYKK 181
> SPAC15E1.06
Length=187
Score = 144 bits (362), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 75/185 (40%), Positives = 108/185 (58%), Gaps = 4/185 (2%)
Query 21 LVLIIGDFHVPQRAVDLPPCFKELLHTDKIRHVLCTGNVGSESVLEFLRGIAGSVHIVKG 80
LVL+IGDFH+P RA L F++LL KI ++C GN+ S SV E+L+ + + +VKG
Sbjct 2 LVLVIGDFHIPDRAPKLSEKFRQLLIPGKISQIICLGNLTSTSVYEYLKHVCSDLKLVKG 61
Query 81 DMDEGMDFPVYKILQFGEFKVALLHGHQVIPWGDADALLQWQRRLDCDIVVSGHTHSNSV 140
D P+ + G FK+ +GH V+P +AL R +D DI++ G TH +
Sbjct 62 AFDISSKAPIAGKITLGSFKIGYTNGHLVVPQDSPEALSILAREMDADILLFGGTHKFAA 121
Query 141 REVEGRFFINPGSATGAYQPWA----PSPTPSFMLMALQGASVVLYIYEERDGKAEVVMS 196
E++G FF+NPGSATGA A PSF+LM +QGA ++LY+Y DG+ V
Sbjct 122 YELDGCFFVNPGSATGAPNVSAVEDDEKIVPSFVLMDVQGAVLILYVYRIFDGEVRVEKM 181
Query 197 EFSKP 201
++ KP
Sbjct 182 QYRKP 186
> CE23493
Length=157
Score = 138 bits (348), Expect = 7e-33, Method: Compositional matrix adjust.
Identities = 64/146 (43%), Positives = 96/146 (65%), Gaps = 2/146 (1%)
Query 50 IRHVLCTGNVGSESVLEFLRGIAGSVHIVKGDMD-EGMDFPVYKILQFGEFKVALLHGHQ 108
++HVLCTGN+ S ++LR ++ VHIV+G+ D E + +P K++ G+F++ + HGHQ
Sbjct 1 MQHVLCTGNLCSRETFDYLRTLSSDVHIVRGEFDDETLKYPDTKVVTVGQFRIGVCHGHQ 60
Query 109 VIPWGDADALLQWQRRLDCDIVVSGHTHSNSVREVEGRFFINPGSATGAYQPWAPSP-TP 167
+IPWGD L R+LD DI+V+G+T+ S E GRFF++PGSATG++ P TP
Sbjct 61 IIPWGDQRMLELLARQLDVDILVTGNTYECSAVEKNGRFFVDPGSATGSFSVTKTEPTTP 120
Query 168 SFMLMALQGASVVLYIYEERDGKAEV 193
SF L+ +Q +VV Y+Y D +V
Sbjct 121 SFALLDVQADNVVTYLYRLIDDAVKV 146
> YHR012w
Length=282
Score = 105 bits (262), Expect = 7e-23, Method: Compositional matrix adjust.
Identities = 68/203 (33%), Positives = 100/203 (49%), Gaps = 38/203 (18%)
Query 21 LVLIIGDFHVPQRAVDLPPCFKELLHT-DKIRHVLCTGN-VGSESVLEFLRGIAGSVHIV 78
L+L + D H+P RA DLP FK+LL DKI V GN S L+F+ I+ ++ IV
Sbjct 2 LLLALSDAHIPDRATDLPVKFKKLLSVPDKISQVALLGNSTKSYDFLKFVNQISNNITIV 61
Query 79 KGDMDEG-----------------MDFPVYKILQFGEFKVALLHGHQVIPWGDADALLQW 121
+G+ D G + P+ I++ G K+ G+ V+P D +LL
Sbjct 62 RGEFDNGHLPSTKKDKASDNSRPMEEIPMNSIIRQGALKIGCCSGYTVVPKNDPLSLLAL 121
Query 122 QRRLDCDIVVSGHTHSNSVREVEGRFFINPGSATGAYQPWAPSPTPSFMLMALQGASVVL 181
R+LD DI++ G TH+ +EG+FF+NPGS TGA+ P ++
Sbjct 122 ARQLDVDILLWGGTHNVEAYTLEGKFFVNPGSCTGAFNTDWP----------------IV 165
Query 182 YIYEERDGKAEVVMSEFSKPAKK 204
+ E+ D E V SE KP K+
Sbjct 166 FDVEDSD---EAVTSEVDKPTKE 185
> CE04536
Length=2214
Score = 29.6 bits (65), Expect = 4.3, Method: Composition-based stats.
Identities = 22/87 (25%), Positives = 32/87 (36%), Gaps = 5/87 (5%)
Query 26 GDFHVPQRAVDLPPCFK-ELLHTDKIRHVLCTGNVGSESVLEFLRGIAGSVHI----VKG 80
GD P ++ LP F LH +++L SE + S+H+ G
Sbjct 159 GDLIFPDESIQLPTMFTANYLHLPPGQYMLGPRADTSEQFCTMMMSSRSSIHVSGGFTSG 218
Query 81 DMDEGMDFPVYKILQFGEFKVALLHGH 107
D E D+P K F V +H
Sbjct 219 DQAERSDYPNLKFTYFDTESVVAIHAQ 245
> Hs17488041
Length=200
Score = 28.9 bits (63), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 18/56 (32%), Positives = 24/56 (42%), Gaps = 0/56 (0%)
Query 122 QRRLDCDIVVSGHTHSNSVREVEGRFFINPGSATGAYQPWAPSPTPSFMLMALQGA 177
Q + CD+ V H + E F+I PGS T +P S L A+ GA
Sbjct 98 QLKTKCDLYVPEHRREVEIGETGLPFYIQPGSDTCDGCEPQQAPPQSLWLFAIPGA 153
> Hs17455610
Length=161
Score = 28.5 bits (62), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 13/32 (40%), Positives = 19/32 (59%), Gaps = 2/32 (6%)
Query 108 QVIPWGDADALLQ--WQRRLDCDIVVSGHTHS 137
Q + W +A ALL+ W C+++V HTHS
Sbjct 84 QPLSWEEAVALLEEFWPSNCVCEVLVHSHTHS 115
Lambda K H
0.323 0.140 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3622842326
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40