bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0001_orf1
Length=109
Score E
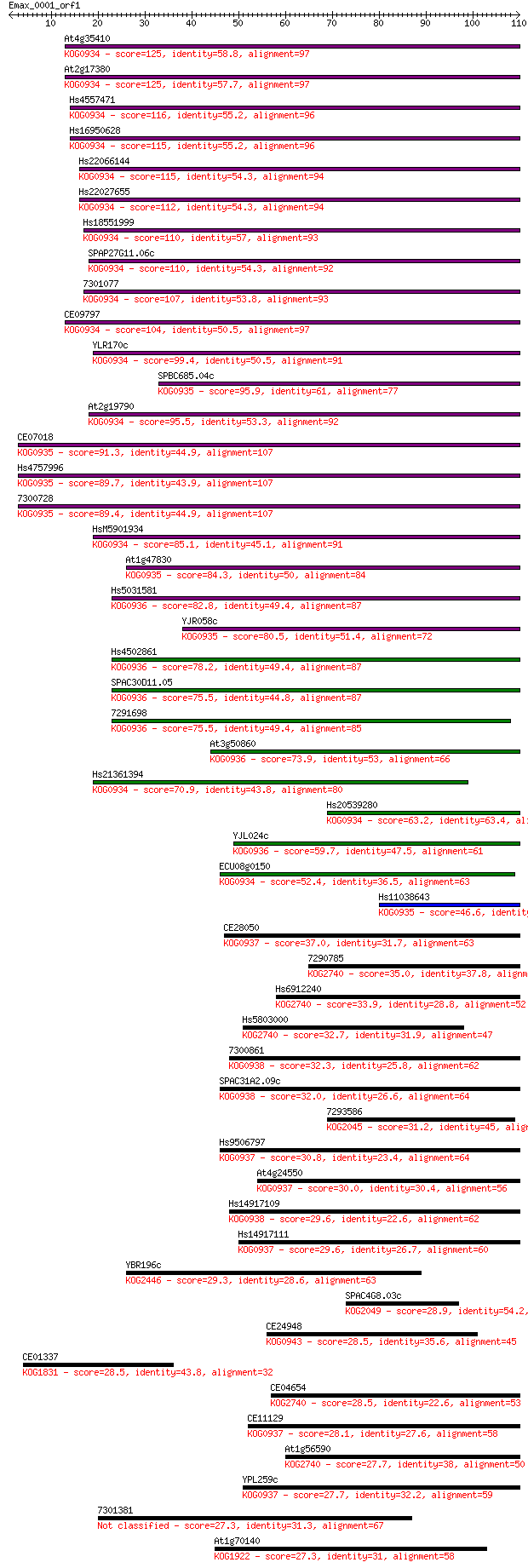
Sequences producing significant alignments: (Bits) Value
At4g35410 125 2e-29
At2g17380 125 2e-29
Hs4557471 116 8e-27
Hs16950628 115 2e-26
Hs22066144 115 3e-26
Hs22027655 112 2e-25
Hs18551999 110 5e-25
SPAP27G11.06c 110 7e-25
7301077 107 4e-24
CE09797 104 3e-23
YLR170c 99.4 1e-21
SPBC685.04c 95.9 2e-20
At2g19790 95.5 2e-20
CE07018 91.3 4e-19
Hs4757996 89.7 1e-18
7300728 89.4 1e-18
HsM5901934 85.1 3e-17
At1g47830 84.3 4e-17
Hs5031581 82.8 2e-16
YJR058c 80.5 7e-16
Hs4502861 78.2 3e-15
SPAC30D11.05 75.5 2e-14
7291698 75.5 2e-14
At3g50860 73.9 6e-14
Hs21361394 70.9 5e-13
Hs20539280 63.2 1e-10
YJL024c 59.7 1e-09
ECU08g0150 52.4 2e-07
Hs11038643 46.6 1e-05
CE28050 37.0 0.009
7290785 35.0 0.032
Hs6912240 33.9 0.074
Hs5803000 32.7 0.16
7300861 32.3 0.24
SPAC31A2.09c 32.0 0.26
7293586 31.2 0.46
Hs9506797 30.8 0.60
At4g24550 30.0 0.99
Hs14917109 29.6 1.6
Hs14917111 29.6 1.6
YBR196c 29.3 1.7
SPAC4G8.03c 28.9 2.2
CE24948 28.5 3.0
CE01337 28.5 3.1
CE04654 28.5 3.6
CE11129 28.1 4.3
At1g56590 27.7 5.3
YPL259c 27.7 5.9
7301381 27.3 6.3
At1g70140 27.3 6.5
> At4g35410
Length=162
Score = 125 bits (314), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 57/97 (58%), Positives = 76/97 (78%), Gaps = 1/97 (1%)
Query 13 SSSNNKERNILIKEAAAKVLQRNARQCNVVEWKGDTKLVYKRYASLYFIGCVCNKENPLF 72
S KER+ +I+E + +L R + CN VEW+G K+VYKRYASLYF C+ ++N L
Sbjct 21 SPYAQKERSKVIRELSGVILNRGPKLCNFVEWRG-YKVVYKRYASLYFCMCIDQEDNELE 79
Query 73 ILELIHHYVEVLDKYFGNVCELDLIFNFHKAHFLLDE 109
+LE+IHHYVE+LD+YFG+VCELDLIFNFHKA+++LDE
Sbjct 80 VLEIIHHYVEILDRYFGSVCELDLIFNFHKAYYILDE 116
> At2g17380
Length=161
Score = 125 bits (313), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 56/97 (57%), Positives = 75/97 (77%), Gaps = 1/97 (1%)
Query 13 SSSNNKERNILIKEAAAKVLQRNARQCNVVEWKGDTKLVYKRYASLYFIGCVCNKENPLF 72
S KER+ +I+E + +L R + CN +EW+G K+VYKRYASLYF C+ +N L
Sbjct 21 SPYTQKERSKVIRELSGVILNRGPKLCNFIEWRG-YKVVYKRYASLYFCMCIDEADNELE 79
Query 73 ILELIHHYVEVLDKYFGNVCELDLIFNFHKAHFLLDE 109
+LE+IHHYVE+LD+YFG+VCELDLIFNFHKA+++LDE
Sbjct 80 VLEIIHHYVEILDRYFGSVCELDLIFNFHKAYYILDE 116
> Hs4557471
Length=158
Score = 116 bits (291), Expect = 8e-27, Method: Compositional matrix adjust.
Identities = 53/96 (55%), Positives = 73/96 (76%), Gaps = 1/96 (1%)
Query 14 SSNNKERNILIKEAAAKVLQRNARQCNVVEWKGDTKLVYKRYASLYFIGCVCNKENPLFI 73
++++KER +++E VL R + C+ +EW+ D K+VYKRYASLYF + ++N L
Sbjct 22 ATSDKERKKMVRELMQVVLARKPKMCSFLEWR-DLKVVYKRYASLYFCCAIEGQDNELIT 80
Query 74 LELIHHYVEVLDKYFGNVCELDLIFNFHKAHFLLDE 109
LELIH YVE+LDKYFG+VCELD+IFNF KA+F+LDE
Sbjct 81 LELIHRYVELLDKYFGSVCELDIIFNFEKAYFILDE 116
> Hs16950628
Length=133
Score = 115 bits (288), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 53/96 (55%), Positives = 73/96 (76%), Gaps = 1/96 (1%)
Query 14 SSNNKERNILIKEAAAKVLQRNARQCNVVEWKGDTKLVYKRYASLYFIGCVCNKENPLFI 73
++++KER +++E VL R + C+ +EW+ D K+VYKRYASLYF + ++N L
Sbjct 22 ATSDKERKKMVRELMQVVLARKPKMCSFLEWR-DLKVVYKRYASLYFCCAIEGQDNELIT 80
Query 74 LELIHHYVEVLDKYFGNVCELDLIFNFHKAHFLLDE 109
LELIH YVE+LDKYFG+VCELD+IFNF KA+F+LDE
Sbjct 81 LELIHRYVELLDKYFGSVCELDIIFNFEKAYFILDE 116
> Hs22066144
Length=269
Score = 115 bits (287), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 51/94 (54%), Positives = 71/94 (75%), Gaps = 1/94 (1%)
Query 16 NNKERNILIKEAAAKVLQRNARQCNVVEWKGDTKLVYKRYASLYFIGCVCNKENPLFILE 75
++KE+ + +E VL R + C+ +EW+ D K+VYKRYASLYF + +++N L LE
Sbjct 132 SDKEKKKITRELVQTVLARKPKMCSFLEWR-DLKIVYKRYASLYFCCAIEDQDNELITLE 190
Query 76 LIHHYVEVLDKYFGNVCELDLIFNFHKAHFLLDE 109
+IH YVE+LDKYFG+VCELD+IFNF KA+F+LDE
Sbjct 191 IIHRYVELLDKYFGSVCELDIIFNFEKAYFILDE 224
> Hs22027655
Length=157
Score = 112 bits (280), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 51/94 (54%), Positives = 71/94 (75%), Gaps = 1/94 (1%)
Query 16 NNKERNILIKEAAAKVLQRNARQCNVVEWKGDTKLVYKRYASLYFIGCVCNKENPLFILE 75
++KE+ + +E VL R + C+ +EW+ D K+VYKRYASLYF + +++N L LE
Sbjct 23 SDKEKKKITRELVQTVLARKPKMCSFLEWR-DLKIVYKRYASLYFCCAIEDQDNELITLE 81
Query 76 LIHHYVEVLDKYFGNVCELDLIFNFHKAHFLLDE 109
+IH YVE+LDKYFG+VCELD+IFNF KA+F+LDE
Sbjct 82 IIHRYVELLDKYFGSVCELDIIFNFEKAYFILDE 115
> Hs18551999
Length=164
Score = 110 bits (276), Expect = 5e-25, Method: Compositional matrix adjust.
Identities = 53/93 (56%), Positives = 69/93 (74%), Gaps = 1/93 (1%)
Query 17 NKERNILIKEAAAKVLQRNARQCNVVEWKGDTKLVYKRYASLYFIGCVCNKENPLFILEL 76
+KER + +E +L R R + V+WK + KLVYKRYASLYF + N++N L LE+
Sbjct 25 DKERKKITREIVQIILSRGHRTSSFVDWK-ELKLVYKRYASLYFCCAIENQDNELLTLEI 83
Query 77 IHHYVEVLDKYFGNVCELDLIFNFHKAHFLLDE 109
+H YVE+LDKYFGNVCELD+IFNF KA+F+LDE
Sbjct 84 VHRYVELLDKYFGNVCELDIIFNFEKAYFILDE 116
> SPAP27G11.06c
Length=162
Score = 110 bits (275), Expect = 7e-25, Method: Compositional matrix adjust.
Identities = 50/92 (54%), Positives = 72/92 (78%), Gaps = 1/92 (1%)
Query 18 KERNILIKEAAAKVLQRNARQCNVVEWKGDTKLVYKRYASLYFIGCVCNKENPLFILELI 77
KER +I++ ++ V+ R + CN VE+KG+ K+VY+RYASL+F+ + +N L ILE+I
Sbjct 27 KERAKIIRDVSSLVITRKPKMCNFVEYKGE-KIVYRRYASLFFVCGIEQDDNELIILEVI 85
Query 78 HHYVEVLDKYFGNVCELDLIFNFHKAHFLLDE 109
H +VE LDKYFGNVCELDLIFNF KA+++++E
Sbjct 86 HKFVECLDKYFGNVCELDLIFNFEKAYYVMEE 117
> 7301077
Length=153
Score = 107 bits (268), Expect = 4e-24, Method: Compositional matrix adjust.
Identities = 50/93 (53%), Positives = 67/93 (72%), Gaps = 1/93 (1%)
Query 17 NKERNILIKEAAAKVLQRNARQCNVVEWKGDTKLVYKRYASLYFIGCVCNKENPLFILEL 76
+K + + +E +L R + C+ +EWK D K+VYKRYASLYF + +N L LE+
Sbjct 21 DKVKKKITRELVTTILARKPKMCSFLEWK-DCKIVYKRYASLYFCCAIEQNDNELLTLEI 79
Query 77 IHHYVEVLDKYFGNVCELDLIFNFHKAHFLLDE 109
IH YVE+LDKYFG+VCELD+IFNF KA+F+LDE
Sbjct 80 IHRYVELLDKYFGSVCELDIIFNFEKAYFILDE 112
> CE09797
Length=157
Score = 104 bits (260), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 49/97 (50%), Positives = 70/97 (72%), Gaps = 1/97 (1%)
Query 13 SSSNNKERNILIKEAAAKVLQRNARQCNVVEWKGDTKLVYKRYASLYFIGCVCNKENPLF 72
++ +K++ + +E ++L R + C +E+K D K+VYKRYASLYF + +N L
Sbjct 21 TAYPDKQKKKICRELITQILARKPKMCAFLEYK-DLKVVYKRYASLYFCCAIEQNDNELI 79
Query 73 ILELIHHYVEVLDKYFGNVCELDLIFNFHKAHFLLDE 109
LE+IH YVE+LDKYFG+VCELD+IFNF KA+F+LDE
Sbjct 80 TLEVIHRYVELLDKYFGSVCELDIIFNFEKAYFILDE 116
> YLR170c
Length=156
Score = 99.4 bits (246), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 46/92 (50%), Positives = 65/92 (70%), Gaps = 2/92 (2%)
Query 19 ERNILIKEAAAKVLQRNARQCNVVEWKGDTKLVYKRYASLYFI-GCVCNKENPLFILELI 77
E+ ++K+ +L R + CN++E+ D K+VYKRYASLYFI G + +N L LE+I
Sbjct 29 EKAKIVKDLTPTILARKPKMCNIIEY-NDHKVVYKRYASLYFIVGMTPDVDNELLTLEII 87
Query 78 HHYVEVLDKYFGNVCELDLIFNFHKAHFLLDE 109
H +VE +D YFGNVCELD+IFNF K + +L+E
Sbjct 88 HRFVETMDTYFGNVCELDIIFNFSKVYDILNE 119
> SPBC685.04c
Length=143
Score = 95.9 bits (237), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 47/78 (60%), Positives = 61/78 (78%), Gaps = 2/78 (2%)
Query 33 QRNAR-QCNVVEWKGDTKLVYKRYASLYFIGCVCNKENPLFILELIHHYVEVLDKYFGNV 91
QRN + Q N +EW+ ++KLVY+RYA LYF CV + +N L ILE+IH +VE+LD +FGNV
Sbjct 41 QRNQKFQANFLEWE-NSKLVYRRYAGLYFCFCVDSTDNDLAILEMIHFFVEILDSFFGNV 99
Query 92 CELDLIFNFHKAHFLLDE 109
CELDLIFNF+K +LDE
Sbjct 100 CELDLIFNFYKVSAILDE 117
> At2g19790
Length=143
Score = 95.5 bits (236), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 49/92 (53%), Positives = 66/92 (71%), Gaps = 1/92 (1%)
Query 18 KERNILIKEAAAKVLQRNARQCNVVEWKGDTKLVYKRYASLYFIGCVCNKENPLFILELI 77
+ER L E K L RN +QC+ VE + + K+VY+RYASL+F+ V + EN L ILE I
Sbjct 27 EERRALEGEIVRKCLARNDQQCSFVEHR-NYKIVYRRYASLFFMVGVDDDENELAILEFI 85
Query 78 HHYVEVLDKYFGNVCELDLIFNFHKAHFLLDE 109
H VE +DK+FGNVCELD++F+ KAHF+L+E
Sbjct 86 HLLVETMDKHFGNVCELDIMFHLEKAHFMLEE 117
> CE07018
Length=142
Score = 91.3 bits (225), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 48/107 (44%), Positives = 72/107 (67%), Gaps = 1/107 (0%)
Query 3 AANSSSSSSSSSSNNKERNILIKEAAAKVLQRNARQCNVVEWKGDTKLVYKRYASLYFIG 62
A + + ++ E+ LI+E A V R+A+ N VE++ + K+VY+RYA LYF
Sbjct 11 AGKTRLAKWYMHFDDDEKQKLIEEVHACVTVRDAKHTNFVEFR-NFKIVYRRYAGLYFCI 69
Query 63 CVCNKENPLFILELIHHYVEVLDKYFGNVCELDLIFNFHKAHFLLDE 109
CV +N L+ LE IH++VEVL++YF NVCELDL+FNF+K + ++DE
Sbjct 70 CVDITDNNLYYLEAIHNFVEVLNEYFHNVCELDLVFNFYKVYTVVDE 116
> Hs4757996
Length=142
Score = 89.7 bits (221), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 47/107 (43%), Positives = 71/107 (66%), Gaps = 1/107 (0%)
Query 3 AANSSSSSSSSSSNNKERNILIKEAAAKVLQRNARQCNVVEWKGDTKLVYKRYASLYFIG 62
A + + ++ E+ LI+E A V R+A+ N VE++ + K++Y+RYA LYF
Sbjct 11 AGKTRLAKWYMQFDDDEKQKLIEEVHAVVTVRDAKHTNFVEFR-NFKIIYRRYAGLYFCI 69
Query 63 CVCNKENPLFILELIHHYVEVLDKYFGNVCELDLIFNFHKAHFLLDE 109
CV +N L LE IH++VEVL++YF NVCELDL+FNF+K + ++DE
Sbjct 70 CVDVNDNKLAYLEGIHNFVEVLNEYFHNVCELDLVFNFYKVYTVVDE 116
> 7300728
Length=142
Score = 89.4 bits (220), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 48/107 (44%), Positives = 72/107 (67%), Gaps = 1/107 (0%)
Query 3 AANSSSSSSSSSSNNKERNILIKEAAAKVLQRNARQCNVVEWKGDTKLVYKRYASLYFIG 62
A + + + ++ E+ LI+E A V R+A+ N VE++ + K+VY+RYA LYF
Sbjct 11 AGKTRLAKWYMNFDDDEKQKLIEEVHAVVTVRDAKHTNFVEFR-NFKIVYRRYAGLYFCI 69
Query 63 CVCNKENPLFILELIHHYVEVLDKYFGNVCELDLIFNFHKAHFLLDE 109
CV +N L LE IH++VEVL++YF NVCELDL+FNF+K + ++DE
Sbjct 70 CVDVNDNNLCYLEAIHNFVEVLNEYFHNVCELDLVFNFYKVYSVVDE 116
> HsM5901934
Length=144
Score = 85.1 bits (209), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 41/91 (45%), Positives = 61/91 (67%), Gaps = 1/91 (1%)
Query 19 ERNILIKEAAAKVLQRNARQCNVVEWKGDTKLVYKRYASLYFIGCVCNKENPLFILELIH 78
+R +L E L R+ QC+ +E+K D KL+Y++YA+L+ + V + EN + I E IH
Sbjct 27 KRTLLETEVIKSCLSRSNEQCSFIEYK-DFKLIYRQYAALFIVVGVNDTENEMAIYEFIH 85
Query 79 HYVEVLDKYFGNVCELDLIFNFHKAHFLLDE 109
++VEVLD+YF V ELD++FN K H +LDE
Sbjct 86 NFVEVLDEYFSRVSELDIMFNLDKVHIILDE 116
> At1g47830
Length=142
Score = 84.3 bits (207), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 42/84 (50%), Positives = 58/84 (69%), Gaps = 1/84 (1%)
Query 26 EAAAKVLQRNARQCNVVEWKGDTKLVYKRYASLYFIGCVCNKENPLFILELIHHYVEVLD 85
E V+ R+A+ N VE++ K++Y+RYA L+F CV +N L LE IH +VE+LD
Sbjct 34 EVHRLVVNRDAKFTNFVEFRTH-KVIYRRYAGLFFSVCVDITDNELAYLESIHLFVEILD 92
Query 86 KYFGNVCELDLIFNFHKAHFLLDE 109
+F NVCELDL+FNFHK + +LDE
Sbjct 93 HFFSNVCELDLVFNFHKVYLILDE 116
> Hs5031581
Length=193
Score = 82.8 bits (203), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 43/92 (46%), Positives = 60/92 (65%), Gaps = 5/92 (5%)
Query 23 LIKEAAAKVLQRNARQCNVVEWKG-----DTKLVYKRYASLYFIGCVCNKENPLFILELI 77
+++E VL+R+ CN +E D KL+Y+ YA+LYF+ CV + E+ L IL+LI
Sbjct 31 IVRETFHLVLKRDDNICNFLEGGSLIGGSDYKLIYRHYATLYFVFCVDSSESELGILDLI 90
Query 78 HHYVEVLDKYFGNVCELDLIFNFHKAHFLLDE 109
+VE LDK F NVCELDLIF+ K H++L E
Sbjct 91 QVFVETLDKCFENVCELDLIFHMDKVHYILQE 122
> YJR058c
Length=147
Score = 80.5 bits (197), Expect = 7e-16, Method: Compositional matrix adjust.
Identities = 37/73 (50%), Positives = 53/73 (72%), Gaps = 2/73 (2%)
Query 38 QCNVVEWKGDTKLVYKRYASLYFI-GCVCNKENPLFILELIHHYVEVLDKYFGNVCELDL 96
Q N VE+ TKL+Y+RYA LYF+ G + P+++ IH +VEVLD +FGNVCELD+
Sbjct 50 QSNFVEFSDSTKLIYRRYAGLYFVMGVDLLDDEPIYLCH-IHLFVEVLDAFFGNVCELDI 108
Query 97 IFNFHKAHFLLDE 109
+FNF+K + ++DE
Sbjct 109 VFNFYKVYMIMDE 121
> Hs4502861
Length=193
Score = 78.2 bits (191), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 43/92 (46%), Positives = 58/92 (63%), Gaps = 5/92 (5%)
Query 23 LIKEAAAKVLQRNARQCNVVEWK-----GDTKLVYKRYASLYFIGCVCNKENPLFILELI 77
+I+E V +R+ CN +E D KL+Y+ YA+LYF+ CV + E+ L IL+LI
Sbjct 31 IIRETFHLVSKRDENVCNFLEGGLLIGGSDNKLIYRHYATLYFVFCVDSSESELGILDLI 90
Query 78 HHYVEVLDKYFGNVCELDLIFNFHKAHFLLDE 109
+VE LDK F NVCELDLIF+ K H +L E
Sbjct 91 QVFVETLDKCFENVCELDLIFHVDKVHNILAE 122
> SPAC30D11.05
Length=165
Score = 75.5 bits (184), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 39/90 (43%), Positives = 55/90 (61%), Gaps = 3/90 (3%)
Query 23 LIKEAAAKVLQRNARQCNVVE---WKGDTKLVYKRYASLYFIGCVCNKENPLFILELIHH 79
LI + A V R CN +E G +++Y++YA+LYF+ V E+ L IL+LI
Sbjct 31 LIGDIYAAVSTRPPTACNFLESNLIAGKNRIIYRQYATLYFVFVVDEGESELGILDLIQV 90
Query 80 YVEVLDKYFGNVCELDLIFNFHKAHFLLDE 109
+VE LD+ F NVCELDL+F F + H +L E
Sbjct 91 FVEALDRCFNNVCELDLVFKFQEIHAILAE 120
> 7291698
Length=210
Score = 75.5 bits (184), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 42/90 (46%), Positives = 56/90 (62%), Gaps = 5/90 (5%)
Query 23 LIKEAAAKVLQRNARQCNVVEWKG-----DTKLVYKRYASLYFIGCVCNKENPLFILELI 77
+IKE V +R+ CN +E D KL+Y+ YA+LYF+ CV + E+ L IL+LI
Sbjct 31 IIKETFQLVSKRDDNVCNFLEGGSLIGGSDYKLIYRHYATLYFVFCVDSSESELGILDLI 90
Query 78 HHYVEVLDKYFGNVCELDLIFNFHKAHFLL 107
+VE LDK F NVCELDLIF+ H +L
Sbjct 91 QVFVETLDKCFENVCELDLIFHADAVHHIL 120
> At3g50860
Length=145
Score = 73.9 bits (180), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 35/66 (53%), Positives = 46/66 (69%), Gaps = 0/66 (0%)
Query 44 WKGDTKLVYKRYASLYFIGCVCNKENPLFILELIHHYVEVLDKYFGNVCELDLIFNFHKA 103
+ D++LVYK YA+LYF+ EN L +L+LI VE LDK F NVCELD++FN+ K
Sbjct 56 FGPDSRLVYKHYATLYFVLVFDGSENELAMLDLIQVLVETLDKCFSNVCELDIVFNYSKM 115
Query 104 HFLLDE 109
H +LDE
Sbjct 116 HAVLDE 121
> Hs21361394
Length=159
Score = 70.9 bits (172), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 35/80 (43%), Positives = 53/80 (66%), Gaps = 1/80 (1%)
Query 19 ERNILIKEAAAKVLQRNARQCNVVEWKGDTKLVYKRYASLYFIGCVCNKENPLFILELIH 78
+R +L E L R+ QC+ +E+K D KL+Y++YA+L+ + V + EN + I E IH
Sbjct 27 KRTLLETEVIKSCLSRSNEQCSFIEYK-DFKLIYRQYAALFIVVGVNDTENEMAIYEFIH 85
Query 79 HYVEVLDKYFGNVCELDLIF 98
++VEVLD+YF V ELD+ F
Sbjct 86 NFVEVLDEYFSRVSELDVSF 105
> Hs20539280
Length=185
Score = 63.2 bits (152), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 26/41 (63%), Positives = 35/41 (85%), Gaps = 0/41 (0%)
Query 69 NPLFILELIHHYVEVLDKYFGNVCELDLIFNFHKAHFLLDE 109
N L LE IHHY+E+L+KYFG+VCELD++FNF + +F+LDE
Sbjct 120 NELTALETIHHYMELLNKYFGSVCELDVVFNFEEVYFILDE 160
> YJL024c
Length=194
Score = 59.7 bits (143), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 29/61 (47%), Positives = 42/61 (68%), Gaps = 0/61 (0%)
Query 49 KLVYKRYASLYFIGCVCNKENPLFILELIHHYVEVLDKYFGNVCELDLIFNFHKAHFLLD 108
+++YK YA+LYF V ++E+ L IL+LI +VE LD+ F V ELDLIFN+ +L+
Sbjct 74 QIIYKNYATLYFTFIVDDQESELAILDLIQTFVESLDRCFTEVNELDLIFNWQTLESVLE 133
Query 109 E 109
E
Sbjct 134 E 134
> ECU08g0150
Length=98
Score = 52.4 bits (124), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 23/63 (36%), Positives = 40/63 (63%), Gaps = 1/63 (1%)
Query 46 GDTKLVYKRYASLYFIGCVCNKENPLFILELIHHYVEVLDKYFGNVCELDLIFNFHKAHF 105
G +V+ R+ ++ F+ V EN ++IL LI++ + + D++F VCEL I+NF + H
Sbjct 13 GKETVVFNRFGNI-FMAFVVENENEMYILSLINNLMSIYDRFFTKVCELHFIYNFKETHI 71
Query 106 LLD 108
+LD
Sbjct 72 ILD 74
> Hs11038643
Length=104
Score = 46.6 bits (109), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 19/30 (63%), Positives = 27/30 (90%), Gaps = 0/30 (0%)
Query 80 YVEVLDKYFGNVCELDLIFNFHKAHFLLDE 109
+VEVL++YF NVCELDL+FNF+K + ++DE
Sbjct 49 FVEVLNEYFHNVCELDLVFNFYKVYTVVDE 78
> CE28050
Length=422
Score = 37.0 bits (84), Expect = 0.009, Method: Composition-based stats.
Identities = 20/63 (31%), Positives = 33/63 (52%), Gaps = 0/63 (0%)
Query 47 DTKLVYKRYASLYFIGCVCNKENPLFILELIHHYVEVLDKYFGNVCELDLIFNFHKAHFL 106
DT V+ ++ ++Y + + N IL ++ VEV +YF +V E + NF + L
Sbjct 54 DTNFVFIKHTNIYLVSACRSNVNVTMILSFLYKCVEVFSEYFKDVEEESVRDNFVVIYEL 113
Query 107 LDE 109
LDE
Sbjct 114 LDE 116
> 7290785
Length=422
Score = 35.0 bits (79), Expect = 0.032, Method: Composition-based stats.
Identities = 17/46 (36%), Positives = 26/46 (56%), Gaps = 1/46 (2%)
Query 65 CNKE-NPLFILELIHHYVEVLDKYFGNVCELDLIFNFHKAHFLLDE 109
C +E PLF++E +H V+ YFG+ E + N+ + LLDE
Sbjct 68 CKQEVPPLFVIEFLHRVVDTFQDYFGDCSESVIKDNYVVVYELLDE 113
> Hs6912240
Length=418
Score = 33.9 bits (76), Expect = 0.074, Method: Composition-based stats.
Identities = 15/52 (28%), Positives = 26/52 (50%), Gaps = 0/52 (0%)
Query 58 LYFIGCVCNKENPLFILELIHHYVEVLDKYFGNVCELDLIFNFHKAHFLLDE 109
L+F+ + + PLF++E +H + YFG E + N + LL+E
Sbjct 64 LFFVSVIQTEVPPLFVIEFLHRVADTFQDYFGECSEAAIKDNVVIVYELLEE 115
> Hs5803000
Length=418
Score = 32.7 bits (73), Expect = 0.16, Method: Composition-based stats.
Identities = 15/47 (31%), Positives = 26/47 (55%), Gaps = 3/47 (6%)
Query 51 VYKRYASLYFIGCVCNKENPLFILELIHHYVEVLDKYFGNVCELDLI 97
VY+ ++F+ + + PLF++E +H V+ YFG VC +I
Sbjct 59 VYRH--KIFFVAVIQTEVPPLFVIEFLHRVVDTFQDYFG-VCSEPVI 102
> 7300861
Length=438
Score = 32.3 bits (72), Expect = 0.24, Method: Composition-based stats.
Identities = 16/62 (25%), Positives = 29/62 (46%), Gaps = 0/62 (0%)
Query 48 TKLVYKRYASLYFIGCVCNKENPLFILELIHHYVEVLDKYFGNVCELDLIFNFHKAHFLL 107
T + + A+++ N + E + +EV+ YFG + E ++ NF + LL
Sbjct 53 TSFFHIKRANIWLAAVTKQNVNAAMVFEFLLKIIEVMQSYFGKISEENIKNNFVLIYELL 112
Query 108 DE 109
DE
Sbjct 113 DE 114
> SPAC31A2.09c
Length=446
Score = 32.0 bits (71), Expect = 0.26, Method: Composition-based stats.
Identities = 17/64 (26%), Positives = 29/64 (45%), Gaps = 0/64 (0%)
Query 46 GDTKLVYKRYASLYFIGCVCNKENPLFILELIHHYVEVLDKYFGNVCELDLIFNFHKAHF 105
G + +Y ++ LY + N + +LE + ++ L YFG + E + N
Sbjct 50 GSSTYIYTKHEDLYVVAITKGNPNVMIVLEFLESLIQDLTHYFGKLNENTVKDNVSFIFE 109
Query 106 LLDE 109
LLDE
Sbjct 110 LLDE 113
> 7293586
Length=1612
Score = 31.2 bits (69), Expect = 0.46, Method: Composition-based stats.
Identities = 18/44 (40%), Positives = 21/44 (47%), Gaps = 4/44 (9%)
Query 69 NPLFILELIHHYVE----VLDKYFGNVCELDLIFNFHKAHFLLD 108
N I EL HHYV VLD Y+ V D + FH F+ D
Sbjct 428 NDELIEELCHHYVNALQWVLDYYYRGVQSWDWYYPFHYTPFISD 471
> Hs9506797
Length=423
Score = 30.8 bits (68), Expect = 0.60, Method: Composition-based stats.
Identities = 15/64 (23%), Positives = 30/64 (46%), Gaps = 0/64 (0%)
Query 46 GDTKLVYKRYASLYFIGCVCNKENPLFILELIHHYVEVLDKYFGNVCELDLIFNFHKAHF 105
G ++ ++++LY + N + ++ +EV +YF + E + NF +
Sbjct 53 GQVHFLWIKHSNLYLVATTSKNANASLVYSFLYKTIEVFCEYFKELEEESIRDNFVIVYE 112
Query 106 LLDE 109
LLDE
Sbjct 113 LLDE 116
> At4g24550
Length=451
Score = 30.0 bits (66), Expect = 0.99, Method: Composition-based stats.
Identities = 17/56 (30%), Positives = 25/56 (44%), Gaps = 0/56 (0%)
Query 54 RYASLYFIGCVCNKENPLFILELIHHYVEVLDKYFGNVCELDLIFNFHKAHFLLDE 109
+ LYF+ +P +LEL+ V+ Y G + E NF + LLDE
Sbjct 63 KVVGLYFVATTRVNVSPSLVLELLQRIARVIKDYLGVLNEDSFRKNFVLVYELLDE 118
> Hs14917109
Length=435
Score = 29.6 bits (65), Expect = 1.6, Method: Composition-based stats.
Identities = 14/62 (22%), Positives = 29/62 (46%), Gaps = 0/62 (0%)
Query 48 TKLVYKRYASLYFIGCVCNKENPLFILELIHHYVEVLDKYFGNVCELDLIFNFHKAHFLL 107
T + + ++++ N + E ++ +V+ YFG + E ++ NF + LL
Sbjct 53 TSFFHVKRSNIWLAAVTKQNVNAAMVFEFLYKMCDVMAAYFGKISEENIKNNFVLIYELL 112
Query 108 DE 109
DE
Sbjct 113 DE 114
> Hs14917111
Length=453
Score = 29.6 bits (65), Expect = 1.6, Method: Composition-based stats.
Identities = 16/60 (26%), Positives = 29/60 (48%), Gaps = 0/60 (0%)
Query 50 LVYKRYASLYFIGCVCNKENPLFILELIHHYVEVLDKYFGNVCELDLIFNFHKAHFLLDE 109
++ R++ LY + +P +LEL+ +L Y G++ E + N + LLDE
Sbjct 56 FIHIRHSGLYLVVTTSENVSPFSLLELLSRLATLLGDYCGSLGEGTISRNVALVYELLDE 115
> YBR196c
Length=554
Score = 29.3 bits (64), Expect = 1.7, Method: Composition-based stats.
Identities = 18/63 (28%), Positives = 30/63 (47%), Gaps = 3/63 (4%)
Query 26 EAAAKVLQRNARQCNVVEWKGDTKLVYKRYASLYFIGCVCNKENPLFILELIHHYVEVLD 85
++ K ++ + Q EWKG T K+ + IG + P+ + E + HY VLD
Sbjct 132 DSVLKHMKEFSEQVRSGEWKGYTG---KKITDVVNIGIGGSDLGPVMVTEALKHYAGVLD 188
Query 86 KYF 88
+F
Sbjct 189 VHF 191
> SPAC4G8.03c
Length=780
Score = 28.9 bits (63), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 13/26 (50%), Positives = 20/26 (76%), Gaps = 2/26 (7%)
Query 73 ILEL-IHHYVE-VLDKYFGNVCELDL 96
+LEL I Y E +++K+FGN+C+L L
Sbjct 664 VLELNIQPYTERIIEKFFGNICKLSL 689
> CE24948
Length=2944
Score = 28.5 bits (62), Expect = 3.0, Method: Composition-based stats.
Identities = 16/48 (33%), Positives = 24/48 (50%), Gaps = 3/48 (6%)
Query 56 ASLYFIGCVCNKENPLFILELIHHYVEVLDKYFGNVCEL---DLIFNF 100
A YF CN E P+ + +LI +Y + ++F CE DL+ F
Sbjct 2105 ADAYFHFLSCNVERPIELKQLIDYYEDSQRRHFREKCEQTDNDLMIGF 2152
> CE01337
Length=2500
Score = 28.5 bits (62), Expect = 3.1, Method: Composition-based stats.
Identities = 14/32 (43%), Positives = 21/32 (65%), Gaps = 0/32 (0%)
Query 4 ANSSSSSSSSSSNNKERNILIKEAAAKVLQRN 35
AN+ SSS S++ N E +I++AAA + Q N
Sbjct 1548 ANAFSSSLRSTAANPEMKQMIEDAAATITQDN 1579
> CE04654
Length=332
Score = 28.5 bits (62), Expect = 3.6, Method: Composition-based stats.
Identities = 12/53 (22%), Positives = 25/53 (47%), Gaps = 0/53 (0%)
Query 57 SLYFIGCVCNKENPLFILELIHHYVEVLDKYFGNVCELDLIFNFHKAHFLLDE 109
+L+ + + + PL ++E +H ++ +YF + + N LLDE
Sbjct 63 NLFLVAVITVETPPLMVIEFLHRVIQTFTQYFDEFSDSSMKENCVMVFELLDE 115
> CE11129
Length=470
Score = 28.1 bits (61), Expect = 4.3, Method: Composition-based stats.
Identities = 16/58 (27%), Positives = 28/58 (48%), Gaps = 0/58 (0%)
Query 52 YKRYASLYFIGCVCNKENPLFILELIHHYVEVLDKYFGNVCELDLIFNFHKAHFLLDE 109
Y +Y ++Y + N + +L ++ VEV +YF + E + NF + L DE
Sbjct 59 YIKYMNVYLVTISKKNTNVILVLSALYKIVEVFCEYFKTLEEEAVRDNFVIIYELFDE 116
> At1g56590
Length=417
Score = 27.7 bits (60), Expect = 5.3, Method: Composition-based stats.
Identities = 19/51 (37%), Positives = 27/51 (52%), Gaps = 2/51 (3%)
Query 60 FIGCVCNKENPLFILELIHHYVEVLDKYFGNVCELDLIF-NFHKAHFLLDE 109
F+ C + PL +E + +VL +Y G + E DLI NF + LLDE
Sbjct 67 FLACSQVEMPPLMAIEFLCRVADVLSEYLGGLNE-DLIKDNFIIVYELLDE 116
> YPL259c
Length=475
Score = 27.7 bits (60), Expect = 5.9, Method: Composition-based stats.
Identities = 19/60 (31%), Positives = 28/60 (46%), Gaps = 1/60 (1%)
Query 51 VYKRYASLYFIGCVCN-KENPLFILELIHHYVEVLDKYFGNVCELDLIFNFHKAHFLLDE 109
++ ++ LY + V + N I +H VEVL Y V E + NF + LLDE
Sbjct 58 LFIQHNDLYVVAIVTSLSANAAAIFTFLHKLVEVLSDYLKTVEEESIRDNFVIIYELLDE 117
> 7301381
Length=541
Score = 27.3 bits (59), Expect = 6.3, Method: Composition-based stats.
Identities = 21/70 (30%), Positives = 35/70 (50%), Gaps = 4/70 (5%)
Query 20 RNILIKEAAAKVLQRNARQCNVVEWKGDTKLVYKRYA---SLYFIGCVCNKENPLFILEL 76
R + +AA+ VL+ N + N + D K + RY S YF+GC+ + N L
Sbjct 283 RKLAKMQAASAVLENNLLENNFSLTEYD-KGFFNRYTESFSAYFLGCLKSCANYLKTQAG 341
Query 77 IHHYVEVLDK 86
H+ ++LD+
Sbjct 342 YEHHAKLLDE 351
> At1g70140
Length=760
Score = 27.3 bits (59), Expect = 6.5, Method: Composition-based stats.
Identities = 18/61 (29%), Positives = 31/61 (50%), Gaps = 8/61 (13%)
Query 45 KGDTKLVY---KRYASLYFIGCVCNKENPLFILELIHHYVEVLDKYFGNVCELDLIFNFH 101
KG+ + V KR Y G V +NPL + ++ ++ ++DK VC LD++ N
Sbjct 664 KGEERKVMELVKRTTDYYQAGAVTKGKNPLHLFVIVRDFLAMVDK----VC-LDIMRNMQ 718
Query 102 K 102
+
Sbjct 719 R 719
Lambda K H
0.320 0.133 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1199474506
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40