bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: eggV2
2,483,276 sequences; 915,453,621 total letters
Query= Emax_1190_orf4
Length=749
Score E
Sequences producing significant alignments: (Bits) Value
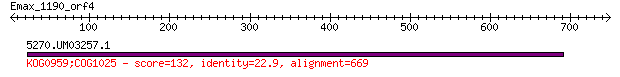
5270.UM03257.1 132 5e-29
> 5270.UM03257.1
Length=1292
Score = 132 bits (332), Expect = 5e-29, Method: Compositional matrix adjust.
Identities = 153/694 (22%), Positives = 300/694 (43%), Gaps = 50/694 (7%)
Query 22 LWVAFGLPATLTSYKKQPTSVLTYLLEYTGEGSLAKRLRLLGLADGISPAV--DRNSIST 79
L +AF +P ++ +P L++ + + GEGS+ L+ G D +S D N
Sbjct 474 LKIAFPIPDQGPHFRSKPGHFLSHFIGHEGEGSILSHLKKKGWCDRLSAGATGDANGFE- 532
Query 80 LLGIKVDLTQKGAAHRGLVLQEIFSYINFLRDHGVGHELVSTMAQQSHIDFHTTQPSSSI 139
I +DLTQ+G + V++ +F YI+ LR + +AQ S + F +
Sbjct 533 FFKISIDLTQEGLDNHEKVVEAVFKYIHLLRSSNLEQWTHDEVAQLSELMFRFKE-KIDP 591
Query 140 MDEAARLAHNL-LTYEPYHVVAGDSLLIDADPRLTNQLLQEMSPSKA---IIAFSDPDFT 195
D A+ A + + Y +++G L D D L Q L ++P ++A + PD +
Sbjct 592 ADYASSTATQMQMPYPREWILSGGWLTRDFDRELITQTLDHLTPQNCRVVVMAKTLPDGS 651
Query 196 SKVDSFETDPYYGVQFRVLDLPQHHAVAMAVLTASPNAFRMPPSLMHIPKASELKILPGL 255
+ +S E +YG ++ + LPQ +LT +P F +H+P+ + +P
Sbjct 652 TSWESKEK--WYGTEYSIKPLPQQ------LLTQTPADFED----LHLPRPNSF--IPVN 697
Query 256 LGLTEPELISEQG-----------GNAGTAVWWQGQGAFAIPRITVQLNGSILKD---KA 301
P L QG N VW + F +P+ V +L++ A
Sbjct 698 FDFKGP-LAEAQGKKPTPRPQLVLDNESIRVWHKLDDRFGLPKANVFF---VLRNPLINA 753
Query 302 DLLSRTQGSLALAAIAEHLQEETVDFQNCGVTHSLAFKGTGFHMGFEAYTEGQLSKLMEH 361
L+ + + + I++ L E + D G+++ L + + Y + ++ L
Sbjct 754 TPLTSIKTRMLIELISDSLVEYSYDASLAGLSYMLDSQDQSLALSLSGYND-KIPVLARS 812
Query 362 VAKLLSDPSMVEPERFERIKQKQMKLVADPATSMAFEHALEAAAILTRNDAFSRKDLLNA 421
+ + L++ V+P RFE +K + + + A + HA L + ++ ++ L
Sbjct 813 ILEKLAN-FQVDPRRFELVKDRVKRSYQNFAIEEPYRHATFYTTYLLQEKMWTPQEKLCE 871
Query 422 LQQTNYDDSFAKLNEL-KNVHVDAFVMGNIDRDQSLTMVEEFLEQAGFTPIDHDDAVASL 480
L+Q N D+ L +L + +H++ GN+ +++++ + P++ + ++S
Sbjct 872 LEQLNVDEVQQFLPDLLQRMHLEVLAHGNLAKEEAIELSNMAWNTIKSRPVNKTELLSSR 931
Query 481 AMEQKQTIEATLANPIKGDKDHASLVQF--QLGIPS-IEDRVNLAVLTQFLNRRIYDSLR 537
++ + P+ + S +++ Q+G P+ +E R L++ +Q N ++D LR
Sbjct 932 SLLLPEKSNKIWNLPVTNAANVNSAIEYYVQIGEPTDVEMRATLSLFSQIANEPVFDQLR 991
Query 538 TEAQLGYIAGAKESQAASTALLQCFVEGAKAHPDEVVKMIDEELSKAKEYLANMPEAEM- 596
T+ QLGY+ + ++ + + V+ + P + +D L + + L M E E
Sbjct 992 TKEQLGYLVFSGIRRSTGSLGWRVIVQSERDAP-YLEGRVDAFLDQFRATLDKMTEQEFE 1050
Query 597 ARWKEAAHAKLTKMEANFSEDFKKSAEEIFAHSNCFTKRDLEVKYLDNDFSRKQLLRTFA 656
A + H KL ++ N E+ + +F + F R +V+ + +++Q++ F
Sbjct 1051 AHKRSIIHKKLENVK-NLVEESTRFWSPVFGGNYDFLARYADVEAIAQT-TKEQVVDLFM 1108
Query 657 KLSDPSRRMVVKLIADLEPEKEVTLIGEAKAQDA 690
K PS KL L L A DA
Sbjct 1109 KYIHPSSPTRSKLSVHLNSTASPALRFSTNAVDA 1142
Lambda K H
0.317 0.131 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 354147316105
Database: eggV2
Posted date: Dec 15, 2009 4:47 PM
Number of letters in database: 915,453,621
Number of sequences in database: 2,483,276
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40