bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_3268_orf1
Length=72
Score E
Sequences producing significant alignments: (Bits) Value
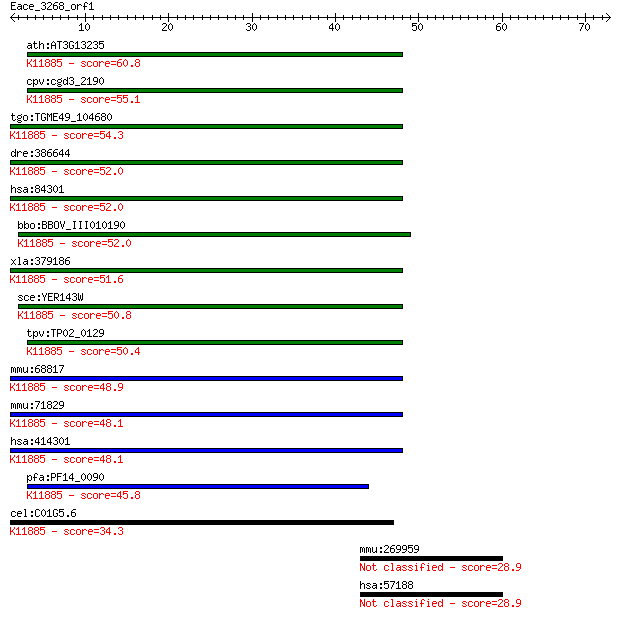
ath:AT3G13235 ubiquitin family protein; K11885 DNA damage-indu... 60.8 1e-09
cpv:cgd3_2190 ubiquitin domain containing protein with a UBA d... 55.1 6e-08
tgo:TGME49_104680 DNA-damage inducible protein, putative ; K11... 54.3 1e-07
dre:386644 ddi2, zgc:63515; DNA-damage inducible protein 2; K1... 52.0 4e-07
hsa:84301 DDI2, MGC14844, RP4-680D5.5; DNA-damage inducible 1 ... 52.0 4e-07
bbo:BBOV_III010190 17.m07883; hypothetical protein; K11885 DNA... 52.0 5e-07
xla:379186 ddi2, MGC53726; protein DDI1 homolog 2; K11885 DNA ... 51.6 6e-07
sce:YER143W DDI1, VSM1; DNA damage-inducible V-SNARE binding p... 50.8 1e-06
tpv:TP02_0129 hypothetical protein; K11885 DNA damage-inducibl... 50.4 1e-06
mmu:68817 Ddi2, 1110056G13Rik, 1700027M01Rik, 9130022E05Rik, A... 48.9 4e-06
mmu:71829 Ddi1, 1700011N24Rik; DDI1, DNA-damage inducible 1, h... 48.1 6e-06
hsa:414301 DDI1, FLJ36017; DNA-damage inducible 1 homolog 1 (S... 48.1 7e-06
pfa:PF14_0090 DNA-damage inducible protein, putative; K11885 D... 45.8 3e-05
cel:C01G5.6 hypothetical protein; K11885 DNA damage-inducible ... 34.3 0.11
mmu:269959 Adamtsl3, 9230119C12Rik, C130057K09, KIAA1233, mKIA... 28.9 3.7
hsa:57188 ADAMTSL3, KIAA1233, MGC150716, MGC150717; ADAMTS-like 3 28.9
> ath:AT3G13235 ubiquitin family protein; K11885 DNA damage-inducible
protein 1
Length=414
Score = 60.8 bits (146), Expect = 1e-09, Method: Composition-based stats.
Identities = 22/45 (48%), Positives = 34/45 (75%), Gaps = 0/45 (0%)
Query 3 ADKCSLASIIDKRFAGLTMGVGKAPIMGRVHVAPLKLGTKFCPCS 47
A++C L ++D+R+ G+ GVG+ I+GR+HVAP+K+G F PCS
Sbjct 231 AERCGLLRLMDQRYKGIAHGVGQTEILGRIHVAPIKIGNNFYPCS 275
> cpv:cgd3_2190 ubiquitin domain containing protein with a UBA
domain at the C-terminus ; K11885 DNA damage-inducible protein
1
Length=384
Score = 55.1 bits (131), Expect = 6e-08, Method: Composition-based stats.
Identities = 22/45 (48%), Positives = 33/45 (73%), Gaps = 0/45 (0%)
Query 3 ADKCSLASIIDKRFAGLTMGVGKAPIMGRVHVAPLKLGTKFCPCS 47
A+KC+L +ID RF+G+ GVG + I+G++HVA +K+G F P S
Sbjct 233 AEKCNLVRLIDYRFSGIAQGVGTSKIVGKIHVAQMKIGNSFFPFS 277
> tgo:TGME49_104680 DNA-damage inducible protein, putative ; K11885
DNA damage-inducible protein 1
Length=527
Score = 54.3 bits (129), Expect = 1e-07, Method: Composition-based stats.
Identities = 23/47 (48%), Positives = 33/47 (70%), Gaps = 0/47 (0%)
Query 1 AFADKCSLASIIDKRFAGLTMGVGKAPIMGRVHVAPLKLGTKFCPCS 47
A A KCSL ++D R+ G+ GVGK I+G++H+A LK+G +F P S
Sbjct 326 ACAQKCSLLRLMDTRYRGVAQGVGKTEIVGKIHLATLKIGQRFFPSS 372
> dre:386644 ddi2, zgc:63515; DNA-damage inducible protein 2;
K11885 DNA damage-inducible protein 1
Length=409
Score = 52.0 bits (123), Expect = 4e-07, Method: Composition-based stats.
Identities = 20/47 (42%), Positives = 34/47 (72%), Gaps = 0/47 (0%)
Query 1 AFADKCSLASIIDKRFAGLTMGVGKAPIMGRVHVAPLKLGTKFCPCS 47
A A++C++ ++D+R+AG+ GVG I+GRVH+A +++ F PCS
Sbjct 273 ACAERCNIMRLVDRRWAGIAKGVGTQKIIGRVHLAQVQIEGDFLPCS 319
> hsa:84301 DDI2, MGC14844, RP4-680D5.5; DNA-damage inducible
1 homolog 2 (S. cerevisiae); K11885 DNA damage-inducible protein
1
Length=399
Score = 52.0 bits (123), Expect = 4e-07, Method: Composition-based stats.
Identities = 20/47 (42%), Positives = 34/47 (72%), Gaps = 0/47 (0%)
Query 1 AFADKCSLASIIDKRFAGLTMGVGKAPIMGRVHVAPLKLGTKFCPCS 47
A A++C++ ++D+R+AG+ GVG I+GRVH+A +++ F PCS
Sbjct 263 ACAERCNIMRLVDRRWAGIAKGVGTQKIIGRVHLAQVQIEGDFLPCS 309
> bbo:BBOV_III010190 17.m07883; hypothetical protein; K11885 DNA
damage-inducible protein 1
Length=500
Score = 52.0 bits (123), Expect = 5e-07, Method: Composition-based stats.
Identities = 21/47 (44%), Positives = 35/47 (74%), Gaps = 0/47 (0%)
Query 2 FADKCSLASIIDKRFAGLTMGVGKAPIMGRVHVAPLKLGTKFCPCSS 48
+A+KC+L +IID+RF G+ +G+ K I+G++H+A +K+G F SS
Sbjct 353 YAEKCNLLNIIDRRFQGVAVGISKERIIGKIHMAQMKIGNLFLLFSS 399
> xla:379186 ddi2, MGC53726; protein DDI1 homolog 2; K11885 DNA
damage-inducible protein 1
Length=393
Score = 51.6 bits (122), Expect = 6e-07, Method: Composition-based stats.
Identities = 20/47 (42%), Positives = 33/47 (70%), Gaps = 0/47 (0%)
Query 1 AFADKCSLASIIDKRFAGLTMGVGKAPIMGRVHVAPLKLGTKFCPCS 47
A A++C + ++D+R+AG+ GVG I+GRVH+A +++ F PCS
Sbjct 257 ACAERCHIMRLVDRRWAGIAKGVGTQKIIGRVHLAQVQIEGDFLPCS 303
> sce:YER143W DDI1, VSM1; DNA damage-inducible V-SNARE binding
protein, contains a ubiquitin-associated (UBA) domain, may
act as a negative regulator of constitutive exocytosis, may
play a role in S-phase checkpoint control; K11885 DNA damage-inducible
protein 1
Length=428
Score = 50.8 bits (120), Expect = 1e-06, Method: Composition-based stats.
Identities = 22/46 (47%), Positives = 30/46 (65%), Gaps = 0/46 (0%)
Query 2 FADKCSLASIIDKRFAGLTMGVGKAPIMGRVHVAPLKLGTKFCPCS 47
A K L+ +IDKRF G GVG I+GR+H A +K+ T++ PCS
Sbjct 232 LAKKTGLSRMIDKRFIGEARGVGTGKIIGRIHQAQVKIETQYIPCS 277
> tpv:TP02_0129 hypothetical protein; K11885 DNA damage-inducible
protein 1
Length=359
Score = 50.4 bits (119), Expect = 1e-06, Method: Composition-based stats.
Identities = 19/45 (42%), Positives = 33/45 (73%), Gaps = 0/45 (0%)
Query 3 ADKCSLASIIDKRFAGLTMGVGKAPIMGRVHVAPLKLGTKFCPCS 47
A +C+L S++D+RF G+ +GVG +G++H+A +K+G+ F P S
Sbjct 228 ALQCNLLSLVDERFKGVAVGVGSTKTLGKIHLADMKIGSIFIPVS 272
> mmu:68817 Ddi2, 1110056G13Rik, 1700027M01Rik, 9130022E05Rik,
AI604911, AU040698; DNA-damage inducible protein 2; K11885
DNA damage-inducible protein 1
Length=399
Score = 48.9 bits (115), Expect = 4e-06, Method: Composition-based stats.
Identities = 19/47 (40%), Positives = 33/47 (70%), Gaps = 0/47 (0%)
Query 1 AFADKCSLASIIDKRFAGLTMGVGKAPIMGRVHVAPLKLGTKFCPCS 47
A A++C++ ++D+R+AG+ GVG I+GRVH+A +++ F CS
Sbjct 263 ACAERCNIMRLVDRRWAGIAKGVGTQKIIGRVHLAQVQIEGDFLACS 309
> mmu:71829 Ddi1, 1700011N24Rik; DDI1, DNA-damage inducible 1,
homolog 1 (S. cerevisiae); K11885 DNA damage-inducible protein
1
Length=408
Score = 48.1 bits (113), Expect = 6e-06, Method: Composition-based stats.
Identities = 19/47 (40%), Positives = 32/47 (68%), Gaps = 0/47 (0%)
Query 1 AFADKCSLASIIDKRFAGLTMGVGKAPIMGRVHVAPLKLGTKFCPCS 47
A A++C++ ++D+R+ G+ GVG IMGRVH+A +++ F CS
Sbjct 277 ACAERCNIMRLVDRRWGGVAKGVGTQRIMGRVHLAQIQIEGDFLQCS 323
> hsa:414301 DDI1, FLJ36017; DNA-damage inducible 1 homolog 1
(S. cerevisiae); K11885 DNA damage-inducible protein 1
Length=396
Score = 48.1 bits (113), Expect = 7e-06, Method: Composition-based stats.
Identities = 19/47 (40%), Positives = 33/47 (70%), Gaps = 0/47 (0%)
Query 1 AFADKCSLASIIDKRFAGLTMGVGKAPIMGRVHVAPLKLGTKFCPCS 47
A A++C++ ++D+R+AG+ GVG I+GRVH+A +++ F CS
Sbjct 271 ACAERCNIMRLVDRRWAGVAKGVGTQRIIGRVHLAQIQIEGDFLQCS 317
> pfa:PF14_0090 DNA-damage inducible protein, putative; K11885
DNA damage-inducible protein 1
Length=382
Score = 45.8 bits (107), Expect = 3e-05, Method: Composition-based stats.
Identities = 17/41 (41%), Positives = 28/41 (68%), Gaps = 0/41 (0%)
Query 3 ADKCSLASIIDKRFAGLTMGVGKAPIMGRVHVAPLKLGTKF 43
A KC++ ++DKRF G+ GVG I+G++H+ +K+G F
Sbjct 275 AQKCNILRLMDKRFTGIAKGVGTKTILGKIHMIDIKIGNYF 315
> cel:C01G5.6 hypothetical protein; K11885 DNA damage-inducible
protein 1
Length=389
Score = 34.3 bits (77), Expect = 0.11, Method: Composition-based stats.
Identities = 16/47 (34%), Positives = 26/47 (55%), Gaps = 1/47 (2%)
Query 1 AFADKCSLASIIDKRFAGLTMGV-GKAPIMGRVHVAPLKLGTKFCPC 46
A A++C L +ID+RF + GV G I G++H+ +K+ C
Sbjct 272 ACAERCGLNGLIDRRFQSMARGVGGTEKIEGKIHLCDVKVEDAHFSC 318
> mmu:269959 Adamtsl3, 9230119C12Rik, C130057K09, KIAA1233, mKIAA1233;
ADAMTS-like 3
Length=1706
Score = 28.9 bits (63), Expect = 3.7, Method: Composition-based stats.
Identities = 10/17 (58%), Positives = 13/17 (76%), Gaps = 0/17 (0%)
Query 43 FCPCSSYCFGGHQDADA 59
F PC++ C GGHQ+A A
Sbjct 666 FTPCTATCLGGHQEAIA 682
> hsa:57188 ADAMTSL3, KIAA1233, MGC150716, MGC150717; ADAMTS-like
3
Length=1691
Score = 28.9 bits (63), Expect = 4.6, Method: Composition-based stats.
Identities = 10/17 (58%), Positives = 13/17 (76%), Gaps = 0/17 (0%)
Query 43 FCPCSSYCFGGHQDADA 59
F PC++ C GGHQ+A A
Sbjct 653 FTPCTATCVGGHQEAIA 669
Lambda K H
0.324 0.138 0.441
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2016064836
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40