bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_3027_orf1
Length=118
Score E
Sequences producing significant alignments: (Bits) Value
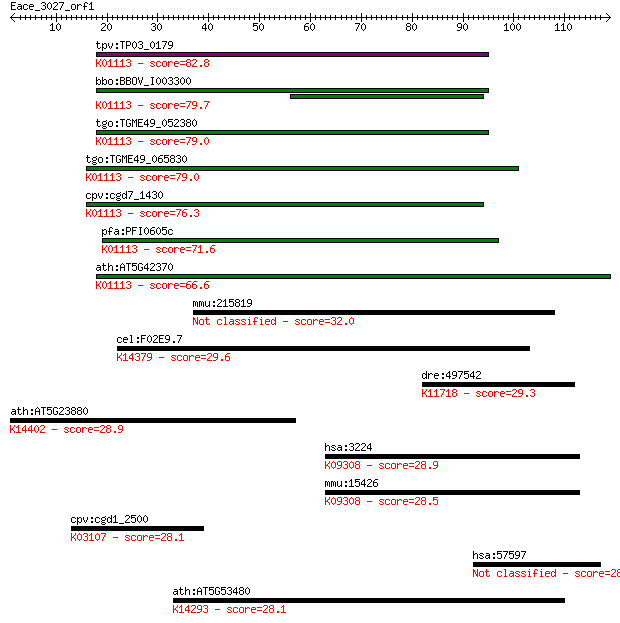
tpv:TP03_0179 hypothetical protein; K01113 alkaline phosphatas... 82.8 3e-16
bbo:BBOV_I003300 19.m02166; hypothetical protein; K01113 alkal... 79.7 2e-15
tgo:TGME49_052380 hypothetical protein ; K01113 alkaline phosp... 79.0 4e-15
tgo:TGME49_065830 hypothetical protein ; K01113 alkaline phosp... 79.0 4e-15
cpv:cgd7_1430 possible phosphodiesterase/alkaline phosphatase ... 76.3 2e-14
pfa:PFI0605c conserved Plasmodium protein, unknown function; K... 71.6 6e-13
ath:AT5G42370 hypothetical protein; K01113 alkaline phosphatas... 66.6 2e-11
mmu:215819 Nhsl1, 5730409E15Rik, A630035H13Rik, A730096F01, BC... 32.0 0.49
cel:F02E9.7 hypothetical protein; K14379 tartrate-resistant ac... 29.6 2.7
dre:497542 uggt2, im:7146988, ugcgl2, wu:fi13a08; UDP-glucose ... 29.3 3.5
ath:AT5G23880 CPSF100 (CLEAVAGE AND POLYADENYLATION SPECIFICIT... 28.9 3.9
hsa:3224 HOXC8, HOX3, HOX3A; homeobox C8; K09308 homeobox prot... 28.9 4.6
mmu:15426 Hoxc8, D130011F21Rik, Hox-3.1; homeobox C8; K09308 h... 28.5 4.7
cpv:cgd1_2500 signal recognition particle SPR68 ; K03107 signa... 28.1 6.2
hsa:57597 BAHCC1, BAHD2, FLJ23058, KIAA1447; BAH domain and co... 28.1 7.3
ath:AT5G53480 importin beta-2, putative; K14293 importin subun... 28.1 7.5
> tpv:TP03_0179 hypothetical protein; K01113 alkaline phosphatase
D [EC:3.1.3.1]
Length=444
Score = 82.8 bits (203), Expect = 3e-16, Method: Composition-based stats.
Identities = 37/77 (48%), Positives = 49/77 (63%), Gaps = 1/77 (1%)
Query 18 GDMLGEQQWAWLQHELQSSTADVNVVVSSLQAFANHRLVTETWSHFPWARGRLFALLAGS 77
D LGE+QW WL+ +L S A +++VSS Q F +R + E+W HFP ++ RL LL +
Sbjct 238 ADALGEEQWKWLESQLVDSEATSHIIVSSFQVFT-YRPMGESWGHFPNSKKRLLDLLEAT 296
Query 78 GAKTPIVLSGDTHFAEF 94
K PI LSGD HFAE
Sbjct 297 KPKKPIFLSGDVHFAEL 313
> bbo:BBOV_I003300 19.m02166; hypothetical protein; K01113 alkaline
phosphatase D [EC:3.1.3.1]
Length=754
Score = 79.7 bits (195), Expect = 2e-15, Method: Composition-based stats.
Identities = 37/77 (48%), Positives = 47/77 (61%), Gaps = 1/77 (1%)
Query 18 GDMLGEQQWAWLQHELQSSTADVNVVVSSLQAFANHRLVTETWSHFPWARGRLFALLAGS 77
GD LG +QW WLQ +L STA+ +++VSS Q F L TE+W P A+ RL LL +
Sbjct 527 GDTLGAEQWKWLQGQLHGSTAEAHIIVSSFQIFTRFPL-TESWGLLPLAKDRLVDLLLTT 585
Query 78 GAKTPIVLSGDTHFAEF 94
K PI LSGD H+ E
Sbjct 586 KPKNPIFLSGDVHYGEL 602
Score = 37.4 bits (85), Expect = 0.012, Method: Composition-based stats.
Identities = 15/38 (39%), Positives = 22/38 (57%), Gaps = 0/38 (0%)
Query 56 VTETWSHFPWARGRLFALLAGSGAKTPIVLSGDTHFAE 93
+TE+W P A+ RL LL + K + +SGD H+ E
Sbjct 187 ITESWGLLPQAKDRLVDLLLAAKPKNTVFVSGDVHWGE 224
> tgo:TGME49_052380 hypothetical protein ; K01113 alkaline phosphatase
D [EC:3.1.3.1]
Length=1222
Score = 79.0 bits (193), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 38/80 (47%), Positives = 50/80 (62%), Gaps = 3/80 (3%)
Query 18 GDMLGEQQWAWLQHELQSSTAD---VNVVVSSLQAFANHRLVTETWSHFPWARGRLFALL 74
GD+LG QQW WL+ EL+ S + V + SS+Q + + TE WS FP AR RL L+
Sbjct 301 GDILGNQQWRWLKEELRKSKEEGDAVTFIASSIQVLPSPFVATEAWSIFPDARRRLLNLI 360
Query 75 AGSGAKTPIVLSGDTHFAEF 94
SG + P++LSGD HFAE
Sbjct 361 MSSGVQIPVLLSGDVHFAEM 380
> tgo:TGME49_065830 hypothetical protein ; K01113 alkaline phosphatase
D [EC:3.1.3.1]
Length=614
Score = 79.0 bits (193), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 39/85 (45%), Positives = 55/85 (64%), Gaps = 1/85 (1%)
Query 16 FLGDMLGEQQWAWLQHELQSSTADVNVVVSSLQAFANHRLVTETWSHFPWARGRLFALLA 75
+ GD+LGE+QW WL+ +L +STA V+++VSS+Q LV E+W HFP A+ RL LL
Sbjct 451 YEGDILGEEQWRWLEAQLTNSTASVHLIVSSIQVSTTLPLV-ESWGHFPRAKQRLIDLLE 509
Query 76 GSGAKTPIVLSGDTHFAEFAGTPRT 100
+ + LSGD HF E +G+ R
Sbjct 510 STKPAGVLFLSGDVHFGEISGSERN 534
> cpv:cgd7_1430 possible phosphodiesterase/alkaline phosphatase
D, of possible plant or bacterial origin ; K01113 alkaline
phosphatase D [EC:3.1.3.1]
Length=463
Score = 76.3 bits (186), Expect = 2e-14, Method: Composition-based stats.
Identities = 34/78 (43%), Positives = 51/78 (65%), Gaps = 1/78 (1%)
Query 16 FLGDMLGEQQWAWLQHELQSSTADVNVVVSSLQAFANHRLVTETWSHFPWARGRLFALLA 75
+ GDMLG++QW WL+ EL +S A N+++SS+Q + +V E W HFP +R RLF+L+
Sbjct 217 YQGDMLGKEQWDWLEKELSNSKALANIIISSIQVTTQYPIV-EGWGHFPKSRERLFSLIK 275
Query 76 GSGAKTPIVLSGDTHFAE 93
+ K LSGD H+ +
Sbjct 276 KTKPKGLFFLSGDVHWGQ 293
> pfa:PFI0605c conserved Plasmodium protein, unknown function;
K01113 alkaline phosphatase D [EC:3.1.3.1]
Length=446
Score = 71.6 bits (174), Expect = 6e-13, Method: Composition-based stats.
Identities = 31/78 (39%), Positives = 47/78 (60%), Gaps = 1/78 (1%)
Query 19 DMLGEQQWAWLQHELQSSTADVNVVVSSLQAFANHRLVTETWSHFPWARGRLFALLAGSG 78
D+LG +QW WL+ EL +S A ++++SS Q F+NH ++ E W P++ RL L+ +
Sbjct 213 DILGNEQWKWLERELTNSNARAHIIISSTQIFSNH-IINENWGLMPYSLRRLRELIKKTK 271
Query 79 AKTPIVLSGDTHFAEFAG 96
K + LSGD HF G
Sbjct 272 PKGLLFLSGDVHFGSIIG 289
> ath:AT5G42370 hypothetical protein; K01113 alkaline phosphatase
D [EC:3.1.3.1]
Length=447
Score = 66.6 bits (161), Expect = 2e-11, Method: Composition-based stats.
Identities = 39/108 (36%), Positives = 55/108 (50%), Gaps = 12/108 (11%)
Query 18 GDMLGEQQWAWLQHELQSSTADVNVVVSSLQAFANHRLVT------ETWSHFPWARGRLF 71
G +LG+ QW WL++EL +++ ++ SS+Q +N T E+W FP R RLF
Sbjct 193 GSILGDTQWDWLENELSGPRSEITIIGSSVQVISNLSATTGPLFYMESWGRFPKERKRLF 252
Query 72 ALLAGSGAKTPIVLSGDTHFAEFAGTPRTGEDDS-GYWYLDCLPPGLI 118
L+A + I +SGD HF E T D S GY D GL+
Sbjct 253 KLIADTKRDGVIFISGDVHFGEI-----TRYDCSVGYPLYDVTSSGLV 295
> mmu:215819 Nhsl1, 5730409E15Rik, A630035H13Rik, A730096F01,
BC013565, D10Bwg0940e, KIAA1357, mKIAA1357; NHS-like 1
Length=1583
Score = 32.0 bits (71), Expect = 0.49, Method: Compositional matrix adjust.
Identities = 24/73 (32%), Positives = 31/73 (42%), Gaps = 5/73 (6%)
Query 37 TADVNVVVSSLQAFANHRLV--TETWSHFPWARGRLFALLAGSGAKTPIVLSGDTHFAEF 94
T DV V+ S++ H+ V SH + G + L +G P LS DT F
Sbjct 266 TEDVKVIPPSMRRIRAHKGVGVAAQMSHLSGSSGNMSVLSDSAGVVFPSRLSNDTGFHSL 325
Query 95 AGTPRTGEDDSGY 107
PRTG S Y
Sbjct 326 ---PRTGPRASTY 335
> cel:F02E9.7 hypothetical protein; K14379 tartrate-resistant
acid phosphatase type 5 [EC:3.1.3.2]
Length=419
Score = 29.6 bits (65), Expect = 2.7, Method: Composition-based stats.
Identities = 25/83 (30%), Positives = 35/83 (42%), Gaps = 11/83 (13%)
Query 22 GEQQWAWLQHELQSSTADVNVVVSSLQAFANHRLVTETWSHFP--WARGRLFALLAGSGA 79
E+QWAWL++ L++S+A ++ + H V SH P R RL LL
Sbjct 242 AEEQWAWLENNLEASSAQYLII-------SGHYPVHSMSSHGPTDCLRQRLDPLLKRFNV 294
Query 80 KTPIVLSGDTHFAEFAGTPRTGE 102
SG H + P GE
Sbjct 295 NA--YFSGHDHSLQHFTFPGYGE 315
> dre:497542 uggt2, im:7146988, ugcgl2, wu:fi13a08; UDP-glucose
glycoprotein glucosyltransferase 2; K11718 UDP-glucose:glycoprotein
glucosyltransferase [EC:2.4.1.-]
Length=1515
Score = 29.3 bits (64), Expect = 3.5, Method: Composition-based stats.
Identities = 13/31 (41%), Positives = 19/31 (61%), Gaps = 1/31 (3%)
Query 82 PIVLSGDTHFAEFAGTPRTGEDDSG-YWYLD 111
P++LS D + +F +P T DDS + YLD
Sbjct 669 PLILSSDRRYLDFTASPGTILDDSAMFLYLD 699
> ath:AT5G23880 CPSF100 (CLEAVAGE AND POLYADENYLATION SPECIFICITY
FACTOR 100); DNA binding / protein binding; K14402 cleavage
and polyadenylation specificity factor subunit 2
Length=739
Score = 28.9 bits (63), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 14/56 (25%), Positives = 29/56 (51%), Gaps = 0/56 (0%)
Query 1 IAAYRSQLANKEYTEFLGDMLGEQQWAWLQHELQSSTADVNVVVSSLQAFANHRLV 56
+ AY+ QL+ K + + LG+ + AW+ E+ + D+ ++ A + H+ V
Sbjct 605 LCAYKVQLSEKLMSNVIFKKLGDSEVAWVDSEVGKTERDMRSLLPMPGAASPHKPV 660
> hsa:3224 HOXC8, HOX3, HOX3A; homeobox C8; K09308 homeobox protein
HoxB/C/D8
Length=242
Score = 28.9 bits (63), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 16/50 (32%), Positives = 21/50 (42%), Gaps = 0/50 (0%)
Query 63 FPWARGRLFALLAGSGAKTPIVLSGDTHFAEFAGTPRTGEDDSGYWYLDC 112
FP + GR AL+ G G P H +F +G +SGY C
Sbjct 28 FPQSVGRSHALVYGPGGSAPGFQHASHHVQDFFHHGTSGISNSGYQQNPC 77
> mmu:15426 Hoxc8, D130011F21Rik, Hox-3.1; homeobox C8; K09308
homeobox protein HoxB/C/D8
Length=242
Score = 28.5 bits (62), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 16/50 (32%), Positives = 21/50 (42%), Gaps = 0/50 (0%)
Query 63 FPWARGRLFALLAGSGAKTPIVLSGDTHFAEFAGTPRTGEDDSGYWYLDC 112
FP + GR AL+ G G P H +F +G +SGY C
Sbjct 28 FPQSVGRSHALVYGPGGSAPGFQHASHHVQDFFHHGTSGISNSGYQQNPC 77
> cpv:cgd1_2500 signal recognition particle SPR68 ; K03107 signal
recognition particle subunit SRP68
Length=632
Score = 28.1 bits (61), Expect = 6.2, Method: Composition-based stats.
Identities = 12/26 (46%), Positives = 19/26 (73%), Gaps = 2/26 (7%)
Query 13 YTEFLGDMLGEQQWAWLQHELQSSTA 38
YT+ + L +++WA + HELQ+STA
Sbjct 408 YTKLI--FLNDERWANISHELQNSTA 431
> hsa:57597 BAHCC1, BAHD2, FLJ23058, KIAA1447; BAH domain and
coiled-coil containing 1
Length=2608
Score = 28.1 bits (61), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 14/30 (46%), Positives = 17/30 (56%), Gaps = 5/30 (16%)
Query 92 AEFAGTPRTGEDDSGYWYL-----DCLPPG 116
A F G P++G D SGY+ L DC PG
Sbjct 478 ASFPGLPKSGLDKSGYFELPTSSQDCARPG 507
> ath:AT5G53480 importin beta-2, putative; K14293 importin subunit
beta-1
Length=870
Score = 28.1 bits (61), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 21/88 (23%), Positives = 34/88 (38%), Gaps = 11/88 (12%)
Query 33 LQSSTAD-VNVVVSSLQAFANHRLVTETWSHFP----WARGRLFALLAGSGAKTPIVLSG 87
L+ +AD + +V++ F + L + +H W GR+F L GS +TPI+
Sbjct 395 LEGPSADKLMAIVNAALTFMLNALTNDPSNHVKDTTAWTLGRIFEFLHGSTIETPIINQA 454
Query 88 DTH------FAEFAGTPRTGEDDSGYWY 109
+ P E G Y
Sbjct 455 NCQQIITVLIQSMNDAPNVAEKACGALY 482
Lambda K H
0.319 0.134 0.431
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2027872200
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40