bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2950_orf1
Length=107
Score E
Sequences producing significant alignments: (Bits) Value
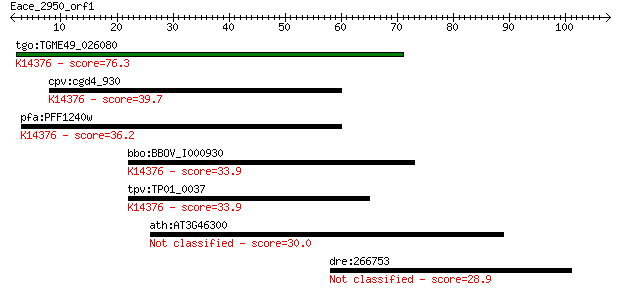
tgo:TGME49_026080 poly(A) polymerase, putative (EC:2.7.7.19); ... 76.3 2e-14
cpv:cgd4_930 pap1p; poly A polymerase (eukaryotic type) ; K143... 39.7 0.002
pfa:PFF1240w poly(A) polymerase PAP, putative (EC:2.7.7.19); K... 36.2 0.026
bbo:BBOV_I000930 16.m00761; poly(A) polymerase PAP; K14376 pol... 33.9 0.12
tpv:TP01_0037 poly(A) polymerase (EC:2.7.7.19); K14376 poly(A)... 33.9 0.12
ath:AT3G46300 hypothetical protein 30.0 1.7
dre:266753 acvrl1, alk1, hm:zehn1109, vgb, zehn1109; activin A... 28.9 3.9
> tgo:TGME49_026080 poly(A) polymerase, putative (EC:2.7.7.19);
K14376 poly(A) polymerase [EC:2.7.7.19]
Length=811
Score = 76.3 bits (186), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 40/69 (57%), Positives = 48/69 (69%), Gaps = 3/69 (4%)
Query 2 SSSMLIALTFASPKAGEAVCDLRPAIAEFVELINGWPERPKLVGSIQLRVRHLRRSQLST 61
+SSML+AL FA V DLRPA EFV+LIN WPER +L G IQLRV+HLRRSQL
Sbjct 660 ASSMLVALNFAQ---QPCVIDLRPAAGEFVDLINQWPERAQLDGQIQLRVKHLRRSQLPA 716
Query 62 EIIKTLKSA 70
++K +A
Sbjct 717 YVLKQKPAA 725
> cpv:cgd4_930 pap1p; poly A polymerase (eukaryotic type) ; K14376
poly(A) polymerase [EC:2.7.7.19]
Length=699
Score = 39.7 bits (91), Expect = 0.002, Method: Composition-based stats.
Identities = 24/53 (45%), Positives = 28/53 (52%), Gaps = 1/53 (1%)
Query 8 ALTFASPKAGEA-VCDLRPAIAEFVELINGWPERPKLVGSIQLRVRHLRRSQL 59
+T S +G A V DLR AI +FV I W E SI L+VRHLR L
Sbjct 461 GVTGVSNSSGTAPVVDLRGAILDFVGFIMNWSEVENYKDSIDLKVRHLRAKDL 513
> pfa:PFF1240w poly(A) polymerase PAP, putative (EC:2.7.7.19);
K14376 poly(A) polymerase [EC:2.7.7.19]
Length=631
Score = 36.2 bits (82), Expect = 0.026, Method: Composition-based stats.
Identities = 17/57 (29%), Positives = 31/57 (54%), Gaps = 0/57 (0%)
Query 3 SSMLIALTFASPKAGEAVCDLRPAIAEFVELINGWPERPKLVGSIQLRVRHLRRSQL 59
SS IAL F E +L AI +F+++ WP++ K + ++ + + +RSQ+
Sbjct 523 SSFFIALIFFYKNPYENTFNLSYAIRDFIDIALNWPQKVKYPNAFKINIMYQKRSQV 579
> bbo:BBOV_I000930 16.m00761; poly(A) polymerase PAP; K14376 poly(A)
polymerase [EC:2.7.7.19]
Length=512
Score = 33.9 bits (76), Expect = 0.12, Method: Composition-based stats.
Identities = 17/51 (33%), Positives = 26/51 (50%), Gaps = 0/51 (0%)
Query 22 DLRPAIAEFVELINGWPERPKLVGSIQLRVRHLRRSQLSTEIIKTLKSAVE 72
D+R +I F E+IN W E I + +R+L+ SQL + K +E
Sbjct 459 DMRMSIKGFKEIINSWTEMEAYKDQIAVNIRYLKNSQLPHFVNPRRKRTIE 509
> tpv:TP01_0037 poly(A) polymerase (EC:2.7.7.19); K14376 poly(A)
polymerase [EC:2.7.7.19]
Length=507
Score = 33.9 bits (76), Expect = 0.12, Method: Composition-based stats.
Identities = 12/43 (27%), Positives = 24/43 (55%), Gaps = 0/43 (0%)
Query 22 DLRPAIAEFVELINGWPERPKLVGSIQLRVRHLRRSQLSTEII 64
D+R ++ F ++IN W + K I ++HL+ +QL ++
Sbjct 452 DIRSSVQSFKDIINNWSDMEKYRDQISFNIKHLKNNQLPDYVL 494
> ath:AT3G46300 hypothetical protein
Length=141
Score = 30.0 bits (66), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 23/65 (35%), Positives = 29/65 (44%), Gaps = 2/65 (3%)
Query 26 AIAEFVELIN--GWPERPKLVGSIQLRVRHLRRSQLSTEIIKTLKSAVEAHQQQRNRSRR 83
A AEFV IN G R +V SI R +LR STE + + R ++
Sbjct 34 AEAEFVRSINHDGSQHRTTVVDSISCRQMYLRSYTFSTEENEEDGDGGDGEAVTRRHNQS 93
Query 84 CRRGG 88
C RGG
Sbjct 94 CFRGG 98
> dre:266753 acvrl1, alk1, hm:zehn1109, vgb, zehn1109; activin
A receptor type II-like 1 (EC:2.7.11.30)
Length=499
Score = 28.9 bits (63), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 14/43 (32%), Positives = 23/43 (53%), Gaps = 0/43 (0%)
Query 58 QLSTEIIKTLKSAVEAHQQQRNRSRRCRRGGKCCLMYLPRWII 100
L TEI+ T AH+ ++R+ +R G+CC+ L +I
Sbjct 304 HLHTEILSTQGKPAIAHRDLKSRNILVKRNGQCCIADLGLAVI 346
Lambda K H
0.324 0.133 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2007980300
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40