bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2938_orf2
Length=154
Score E
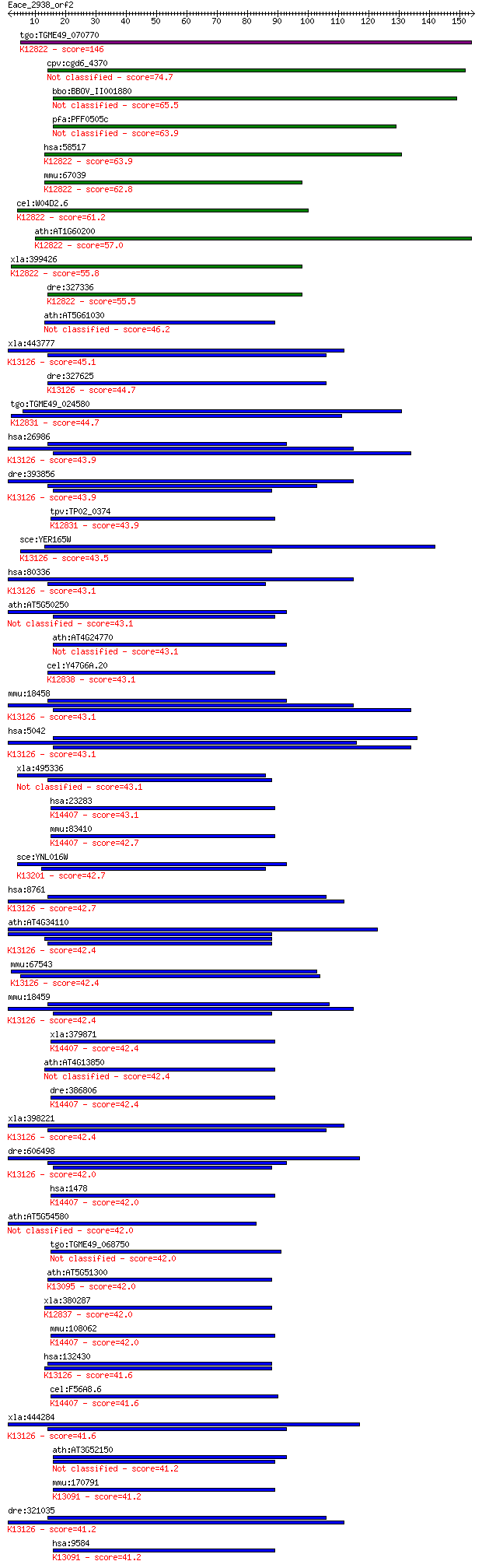
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_070770 hypothetical protein ; K12822 RNA-binding pr... 146 2e-35
cpv:cgd6_4370 SPBC24E9.10-like. RRM domain protein, no PWI domain 74.7 1e-13
bbo:BBOV_II001880 18.m06144; hypothetical protein 65.5 6e-11
pfa:PFF0505c conserved Plasmodium protein, unknown function 63.9 2e-10
hsa:58517 RBM25, MGC105088, MGC117168, NET52, RED120, RNPC7, S... 63.9 2e-10
mmu:67039 Rbm25, 2600011C06Rik, 2610015J01Rik, A130095G20Rik, ... 62.8 4e-10
cel:W04D2.6 hypothetical protein; K12822 RNA-binding protein 25 61.2
ath:AT1G60200 splicing factor PWI domain-containing protein / ... 57.0 2e-08
xla:399426 rbm25, rnpc7; RNA binding motif protein 25; K12822 ... 55.8 5e-08
dre:327336 rbm25, fi03e10, si:ch211-105d18.5, wu:fi03e10, zgc:... 55.5 7e-08
ath:AT5G61030 GR-RBP3 (glycine-rich RNA-binding protein 3); AT... 46.2 4e-05
xla:443777 pabpc1l-b, MGC81363, epab, epabp-b, pabpc1l1; poly(... 45.1 1e-04
dre:327625 pabpc1l, fi20g03, pab-2, wu:fi20g03, zgc:55855; pol... 44.7 1e-04
tgo:TGME49_024580 splicing factor, putative (EC:3.1.2.15); K12... 44.7 1e-04
hsa:26986 PABPC1, PAB1, PABP, PABP1, PABPC2, PABPL1; poly(A) b... 43.9 2e-04
dre:393856 pabpc1b, MGC77608, pabpc1, zgc:77608; poly A bindin... 43.9 2e-04
tpv:TP02_0374 spliceosome associated protein; K12831 splicing ... 43.9 2e-04
sce:YER165W PAB1; Pab1p; K13126 polyadenylate-binding protein 43.5 2e-04
hsa:80336 PABPC1L, C20orf119, FLJ11840, FLJ30809, FLJ42053, PA... 43.1 3e-04
ath:AT5G50250 31 kDa ribonucleoprotein, chloroplast, putative ... 43.1 3e-04
ath:AT4G24770 RBP31; RBP31 (31-KDA RNA BINDING PROTEIN); RNA b... 43.1 3e-04
cel:Y47G6A.20 rnp-6; RNP (RRM RNA binding domain) containing f... 43.1 3e-04
mmu:18458 Pabpc1, PABP, Pabp1, PabpI, Pabpl1, ePAB; poly(A) bi... 43.1 3e-04
hsa:5042 PABPC3, PABP3, PABPL3, tPABP; poly(A) binding protein... 43.1 3e-04
xla:495336 hypothetical LOC495336 43.1 4e-04
hsa:23283 CSTF2T, CstF-64T, DKFZp434C1013, KIAA0689; cleavage ... 43.1 4e-04
mmu:83410 Cstf2t, 64kDa, C77975, tCstF-64, tauCstF-64; cleavag... 42.7 4e-04
sce:YNL016W PUB1, RNP1; Poly (A)+ RNA-binding protein, abundan... 42.7 4e-04
hsa:8761 PABPC4, APP-1, APP1, FLJ43938, PABP4, iPABP; poly(A) ... 42.7 5e-04
ath:AT4G34110 PAB2; PAB2 (POLY(A) BINDING 2); RNA binding / tr... 42.4 6e-04
mmu:67543 Pabpc6, 4932702K14Rik, AI428050, MGC132900, Pabpc3; ... 42.4 6e-04
mmu:18459 Pabpc2, PABP, PABP+, Pabp2; poly(A) binding protein,... 42.4 6e-04
xla:379871 cstf2, MGC53575, MGC83019, cstf-64; cleavage stimul... 42.4 6e-04
ath:AT4G13850 GR-RBP2 (GLYCINE-RICH RNA-BINDING PROTEIN 2); AT... 42.4 7e-04
dre:386806 cstf2, CstF-64, fb11e07, wu:fb11e07, zgc:56346, zgc... 42.4 7e-04
xla:398221 pabpc1l-a, MGC82273, ePAB, epabp-a, pabpc1l1; poly(... 42.4 7e-04
dre:606498 pabpc1a, pabpc1, wu:fb16a02, wu:fi19b08, wu:fj12d09... 42.0 7e-04
hsa:1478 CSTF2, CstF-64; cleavage stimulation factor, 3' pre-R... 42.0 7e-04
ath:AT5G54580 RNA recognition motif (RRM)-containing protein 42.0 8e-04
tgo:TGME49_068750 peptidyl-prolyl cis-trans isomerase E, putative 42.0 8e-04
ath:AT5G51300 splicing factor-related; K13095 splicing factor 1 42.0
xla:380287 u2af2, MGC53441, u2af65; U2 small nuclear RNA auxil... 42.0 9e-04
mmu:108062 Cstf2, 64kDa, C630034J23Rik, Cstf64; cleavage stimu... 42.0 9e-04
hsa:132430 PABPC4L, DKFZp686J06116; poly(A) binding protein, c... 41.6 9e-04
cel:F56A8.6 cpf-2; Cleavage and Polyadenylation Factor family ... 41.6 0.001
xla:444284 pabpc4, MGC80927, PABP, ePAB, ePABP; poly(A) bindin... 41.6 0.001
ath:AT3G52150 RNA recognition motif (RRM)-containing protein 41.2 0.001
mmu:170791 Rbm39, 1500012C14Rik, 2310040E03Rik, B330012G18Rik,... 41.2 0.001
dre:321035 pabpc4, cb12, sb:cb12; poly(A) binding protein, cyt... 41.2 0.001
hsa:9584 RBM39, CAPER, CAPERalpha, CC1.3, DKFZp781C0423, FLJ44... 41.2 0.001
> tgo:TGME49_070770 hypothetical protein ; K12822 RNA-binding
protein 25
Length=779
Score = 146 bits (369), Expect = 2e-35, Method: Compositional matrix adjust.
Identities = 72/149 (48%), Positives = 106/149 (71%), Gaps = 0/149 (0%)
Query 5 QQQQQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAE 64
++ + + T+Y+GNI +H DNDF+R LL E GRV+RWNRQ+DP +GQLAAFGFCDF +
Sbjct 103 EKADESSKPVTVYVGNINRHVDNDFLRLLLAECGRVIRWNRQADPTTGQLAAFGFCDFQD 162
Query 65 PLAAAKCVACLAGRKLHGRSLVVNCNEKVRAKIAKVTEERILATMRAYKNKTREQVEKEI 124
+ A + L + G++L VNCNEKVRA++ +V E+R+L+T+ +++K RE VEKEI
Sbjct 163 AVGALNALEVLPDLVIGGKALKVNCNEKVRAEVNRVKEDRVLSTLHTFRDKKREDVEKEI 222
Query 125 EEEYKRLKDGVQQILDKKNASLPEDDETE 153
EEE RL+ V +++ +K + +P DD E
Sbjct 223 EEETARLRSAVLEVVKRKESQIPPDDAEE 251
> cpv:cgd6_4370 SPBC24E9.10-like. RRM domain protein, no PWI domain.
Length=556
Score = 74.7 bits (182), Expect = 1e-13, Method: Composition-based stats.
Identities = 44/139 (31%), Positives = 69/139 (49%), Gaps = 1/139 (0%)
Query 14 TTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVA 73
TT+Y+G I N +R LL E G +LRW+RQ DP + +L++FG C+F P V+
Sbjct 63 TTVYVGKIPAQVTNTHIRKLLSECGTILRWSRQEDPTTKKLSSFGLCEFDSPEGVINAVS 122
Query 74 CLAGRKLHGRSLVVNCNEKVRAKIAKVTEERILATMRAYKNK-TREQVEKEIEEEYKRLK 132
L G K+ G L+V + +IAK RI ++ T E + E+ +L+
Sbjct 123 ALNGIKISGGQLLVKYQHGIDKEIAKWQSNRINELLKHRGGDCTIETILNELSTSDSKLR 182
Query 133 DGVQQILDKKNASLPEDDE 151
+Q++L L + E
Sbjct 183 ISIQRLLAGMTFELDSEQE 201
> bbo:BBOV_II001880 18.m06144; hypothetical protein
Length=515
Score = 65.5 bits (158), Expect = 6e-11, Method: Composition-based stats.
Identities = 38/148 (25%), Positives = 72/148 (48%), Gaps = 15/148 (10%)
Query 16 LYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVACL 75
+Y+G + +DF+ N++ + G+++ + R +DP SG LA F C+F P A + CL
Sbjct 87 VYVGGLYSGTSSDFVINIMQKCGKLVNFKRHTDPSSGLLANFALCEFETPRGAYYALECL 146
Query 76 AGRKLHGRSLVVNCNEKVRAKIAKVTEERILA---------------TMRAYKNKTREQV 120
A K + V+CN+KVR + +E++ + RA ++ RE++
Sbjct 147 ANLKFGDGEIKVSCNDKVRRLVDAWVKEQMASLREQHGDLNDAELEQLFRAPESDLREEL 206
Query 121 EKEIEEEYKRLKDGVQQILDKKNASLPE 148
E I+ E +++ + + S P+
Sbjct 207 ENLIKYEVMAMQESSSSVGSQPGVSKPD 234
> pfa:PFF0505c conserved Plasmodium protein, unknown function
Length=960
Score = 63.9 bits (154), Expect = 2e-10, Method: Composition-based stats.
Identities = 38/113 (33%), Positives = 65/113 (57%), Gaps = 6/113 (5%)
Query 16 LYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVACL 75
+YIGNI K+ +++ M +L FG V++W RQ +P + +L AFGFC+F++ C+ L
Sbjct 42 MYIGNIDKYIEDNDMVKMLEIFGNVIKWQRQRNPSTNELMAFGFCEFSDIYEVYLCMNIL 101
Query 76 AGRKLHGRSLVVNCNEKVRAKIAKVTEERILATMRAYKNKTREQVEKEIEEEY 128
KL + L VNC++ ++ ERI+ M + + +E+ KE + +Y
Sbjct 102 DNIKLGDKHLKVNCSDNLKKMF-----ERIVDVMYEKREEIKEE-HKERDMQY 148
> hsa:58517 RBM25, MGC105088, MGC117168, NET52, RED120, RNPC7,
S164, Snu71, fSAP94; RNA binding motif protein 25; K12822 RNA-binding
protein 25
Length=843
Score = 63.9 bits (154), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 35/118 (29%), Positives = 64/118 (54%), Gaps = 1/118 (0%)
Query 13 ATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCV 72
TT+++GNI++ A + +R LL + G VL W R SG+L AFGFC++ EP + + +
Sbjct 86 TTTVFVGNISEKASDMLIRQLLAKCGLVLSWKRVQG-ASGKLQAFGFCEYKEPESTLRAL 144
Query 73 ACLAGRKLHGRSLVVNCNEKVRAKIAKVTEERILATMRAYKNKTREQVEKEIEEEYKR 130
L ++ + L+V + K +A++ + ++ + A E+ ++EE KR
Sbjct 145 RLLHDLQIGEKKLLVKVDAKTKAQLDEWKAKKKASNGNARPETVTNDDEEALDEETKR 202
> mmu:67039 Rbm25, 2600011C06Rik, 2610015J01Rik, A130095G20Rik,
AI159652, AL023075, AU043498, RNPC7; RNA binding motif protein
25; K12822 RNA-binding protein 25
Length=838
Score = 62.8 bits (151), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 29/85 (34%), Positives = 51/85 (60%), Gaps = 1/85 (1%)
Query 13 ATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCV 72
TT+++GNI++ A + +R LL + G VL W R SG+L AFGFC++ EP + + +
Sbjct 86 TTTVFVGNISEKASDMLIRQLLAKCGLVLSWKRVQG-ASGKLQAFGFCEYKEPESTLRAL 144
Query 73 ACLAGRKLHGRSLVVNCNEKVRAKI 97
L ++ + L+V + K +A++
Sbjct 145 RLLHDLQIGEKKLLVKVDAKTKAQL 169
> cel:W04D2.6 hypothetical protein; K12822 RNA-binding protein
25
Length=705
Score = 61.2 bits (147), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 31/96 (32%), Positives = 50/96 (52%), Gaps = 1/96 (1%)
Query 4 QQQQQQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFA 63
QQ QQ+ ++ +++GNI+ ++F+R +L E G V W R +G+L FGFC F
Sbjct 37 QQSSQQKPDSSPVFVGNISDKCSDEFIRKILNECGNVASWKRIKGS-NGKLQGFGFCHFT 95
Query 64 EPLAAAKCVACLAGRKLHGRSLVVNCNEKVRAKIAK 99
+ + + L L + L V +EKVR + K
Sbjct 96 DLEGTLRALRILHEFHLGDKKLTVKPDEKVRDDLRK 131
> ath:AT1G60200 splicing factor PWI domain-containing protein
/ RNA recognition motif (RRM)-containing protein; K12822 RNA-binding
protein 25
Length=899
Score = 57.0 bits (136), Expect = 2e-08, Method: Composition-based stats.
Identities = 45/156 (28%), Positives = 77/156 (49%), Gaps = 13/156 (8%)
Query 10 QQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAA 69
++ TT+YIG I +NDFM ++L G V R DP + + FGF +F
Sbjct 200 EKPQTTIYIGKIAT-VENDFMMSILEFCGHVKSCLRAEDPTTKKPKGFGFYEFESAEGIL 258
Query 70 KCVACLAGRKLHGRSLVVNCNEKVRAKIAKVTEERILATMRAYKNK-TREQVEKEIEEEY 128
+ + L R + G+ L+VN N+ + + K E++I +A +++ T+E + E E
Sbjct 259 RAIRLLTQRTIDGQELLVNVNQATKEYLLKYVEKKIETAKKAKESQGTKENQAEGPESEQ 318
Query 129 KRL--------KDGVQQI---LDKKNASLPEDDETE 153
+L KDG +I +D N+++ D+E E
Sbjct 319 DKLESADNETGKDGESKIKENIDIANSAVLTDEERE 354
> xla:399426 rbm25, rnpc7; RNA binding motif protein 25; K12822
RNA-binding protein 25
Length=814
Score = 55.8 bits (133), Expect = 5e-08, Method: Composition-based stats.
Identities = 29/96 (30%), Positives = 55/96 (57%), Gaps = 1/96 (1%)
Query 2 QQQQQQQQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCD 61
+ ++ + TT+++GNI++ A + +R LL + G VL W R SG+L AFGFC+
Sbjct 73 KNKENDENSGPTTTVFVGNISEKASDMLIRQLLAKCGLVLSWKRVQGA-SGKLQAFGFCE 131
Query 62 FAEPLAAAKCVACLAGRKLHGRSLVVNCNEKVRAKI 97
+ EP + + + L ++ + L+V + K +A++
Sbjct 132 YKEPESTLRALRLLHDLQIGEKKLLVKVDAKTKAQL 167
> dre:327336 rbm25, fi03e10, si:ch211-105d18.5, wu:fi03e10, zgc:56208;
RNA binding motif protein 25; K12822 RNA-binding protein
25
Length=731
Score = 55.5 bits (132), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 27/84 (32%), Positives = 50/84 (59%), Gaps = 1/84 (1%)
Query 14 TTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVA 73
TT+++GN+++ A + +R LL + G VL R SG+L AFGFC++ EP + + +
Sbjct 87 TTVFVGNVSEKASDMLIRQLLAKCGLVLSRKRVQG-ASGKLQAFGFCEYKEPESTLRALR 145
Query 74 CLAGRKLHGRSLVVNCNEKVRAKI 97
L ++ + L+V + K +A++
Sbjct 146 LLHELQVGEKKLLVKVDAKTKAQL 169
> ath:AT5G61030 GR-RBP3 (glycine-rich RNA-binding protein 3);
ATP binding / RNA binding
Length=309
Score = 46.2 bits (108), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 25/76 (32%), Positives = 38/76 (50%), Gaps = 0/76 (0%)
Query 13 ATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCV 72
++ L+IG + D D +R ++G V+ D +G+ FGF F AA+ +
Sbjct 39 SSKLFIGGMAYSMDEDSLREAFTKYGEVVDTRVILDRETGRSRGFGFVTFTSSEAASSAI 98
Query 73 ACLAGRKLHGRSLVVN 88
L GR LHGR + VN
Sbjct 99 QALDGRDLHGRVVKVN 114
> xla:443777 pabpc1l-b, MGC81363, epab, epabp-b, pabpc1l1; poly(A)
binding protein, cytoplasmic 1-like; K13126 polyadenylate-binding
protein
Length=629
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 30/113 (26%), Positives = 54/113 (47%), Gaps = 4/113 (3%)
Query 1 QQQQQQQQQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFC 60
+Q +Q++ + + LY+ N+ D+D +R +G + ++ G FGF
Sbjct 281 EQIKQERINRYQGVNLYVKNLDDGIDDDRLRKEFSPYGTITSTKVMTE--GGHSKGFGFV 338
Query 61 DFAEPLAAAKCVACLAGRKLHGRSLVVNCNEKVRAKIAKVTEERI--LATMRA 111
F+ P A K V + GR + + L V ++ + A +T + + LATMRA
Sbjct 339 CFSSPEEATKAVTEMNGRIVSTKPLYVALAQRKEERKAILTNQYMQRLATMRA 391
Score = 38.9 bits (89), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 26/99 (26%), Positives = 47/99 (47%), Gaps = 8/99 (8%)
Query 14 TTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVA 73
T +YI N + D+ ++ + FG L D SG+ FGF ++ A K V
Sbjct 191 TNVYIKNFGEDMDDKRLKEIFSAFGNTLSVKVMMD-NSGRSRGFGFVNYGNHEEAQKAVT 249
Query 74 CLAGRKLHGRSLVVNCNEK-------VRAKIAKVTEERI 105
+ G++++GR + V +K ++ K ++ +ERI
Sbjct 250 EMNGKEVNGRMVYVGRAQKRIERQGELKRKFEQIKQERI 288
> dre:327625 pabpc1l, fi20g03, pab-2, wu:fi20g03, zgc:55855; poly(A)
binding protein, cytoplasmic 1-like; K13126 polyadenylate-binding
protein
Length=620
Score = 44.7 bits (104), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 29/99 (29%), Positives = 51/99 (51%), Gaps = 8/99 (8%)
Query 14 TTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVA 73
T +YI N + D++ ++N+ EFG+ L +D G+ FGF +F A + V
Sbjct 191 TNVYIKNFGEDIDSEKLKNIFTEFGKTLSVCVMTDE-RGRSRGFGFVNFVNHGDARRAVT 249
Query 74 CLAGRKLHGRSLVVNCNEK-------VRAKIAKVTEERI 105
+ G++L+GR L V +K ++ K ++ +ERI
Sbjct 250 EMNGKELNGRVLYVGRAQKRLERQGELKRKFEQIKQERI 288
> tgo:TGME49_024580 splicing factor, putative (EC:3.1.2.15); K12831
splicing factor 3B subunit 4
Length=576
Score = 44.7 bits (104), Expect = 1e-04, Method: Composition-based stats.
Identities = 32/137 (23%), Positives = 59/137 (43%), Gaps = 12/137 (8%)
Query 6 QQQQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEP 65
Q ++ + TLYIGN+ D+D + L + G V + D ++G +GF +F
Sbjct 21 QVYERNQDATLYIGNLDSQVDDDLLWELFVQCGPVRTVSVPRDKLTGNHQGYGFVEFRNE 80
Query 66 LAAAKCVACLAGRKLHGRSLVVNCNEKVRAKI------------AKVTEERILATMRAYK 113
+ A + + KL+G++L +N + + R V E+ I T A+
Sbjct 81 VDADYALKLMNMVKLYGKALRLNKSAQDRRNFDVGANVFLGNLDPDVDEKTIYDTFSAFG 140
Query 114 NKTREQVEKEIEEEYKR 130
N ++ ++ E R
Sbjct 141 NIISAKIMRDPETGLSR 157
Score = 35.0 bits (79), Expect = 0.084, Method: Composition-based stats.
Identities = 25/111 (22%), Positives = 46/111 (41%), Gaps = 2/111 (1%)
Query 2 QQQQQQQQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCD 61
+ Q ++ +++GN+ D + + FG ++ DP +G FGF
Sbjct 104 KSAQDRRNFDVGANVFLGNLDPDVDEKTIYDTFSAFGNIISAKIMRDPETGLSRGFGFVS 163
Query 62 FAEPLAAAKCVACLAGRKLHGRSLVVNCNEK--VRAKIAKVTEERILATMR 110
F A+ +A + G+ + R + V+ K R + ER+LA R
Sbjct 164 FDTFEASDAALAAMNGQFICNRPIHVSYAYKKDTRGERHGSAAERLLAANR 214
> hsa:26986 PABPC1, PAB1, PABP, PABP1, PABPC2, PABPL1; poly(A)
binding protein, cytoplasmic 1; K13126 polyadenylate-binding
protein
Length=636
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 25/79 (31%), Positives = 42/79 (53%), Gaps = 1/79 (1%)
Query 14 TTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVA 73
T +YI N + D++ +++L G+FG L +D SG+ FGF F A K V
Sbjct 191 TNVYIKNFGEDMDDERLKDLFGKFGPALSVKVMTDE-SGKSKGFGFVSFERHEDAQKAVD 249
Query 74 CLAGRKLHGRSLVVNCNEK 92
+ G++L+G+ + V +K
Sbjct 250 EMNGKELNGKQIYVGRAQK 268
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 28/116 (24%), Positives = 54/116 (46%), Gaps = 4/116 (3%)
Query 1 QQQQQQQQQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFC 60
+Q +Q + + + LY+ N+ D++ +R FG + + G+ FGF
Sbjct 281 EQMKQDRITRYQGVNLYVKNLDDGIDDERLRKEFSPFGTITSAKVMME--GGRSKGFGFV 338
Query 61 DFAEPLAAAKCVACLAGRKLHGRSLVVNCNEKVRAKIAKVTEERI--LATMRAYKN 114
F+ P A K V + GR + + L V ++ + A +T + + +A++RA N
Sbjct 339 CFSSPEEATKAVTEMNGRIVATKPLYVALAQRKEERQAHLTNQYMQRMASVRAVPN 394
Score = 30.4 bits (67), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 29/118 (24%), Positives = 51/118 (43%), Gaps = 9/118 (7%)
Query 16 LYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVACL 75
++I N+ K DN + + FG +L D + +GF F AA + + +
Sbjct 101 IFIKNLDKSIDNKALYDTFSAFGNILSCKVVCDENGSK--GYGFVHFETQEAAERAIEKM 158
Query 76 AGRKLHGRSLVVNCNEKVRAKIAKVTEERILATMRAYKNKTREQVEKEIEEEYKRLKD 133
G L+ R + V R K K E + A + + N + +++++E RLKD
Sbjct 159 NGMLLNDRKVFVG-----RFKSRKEREAELGARAKEFTNVYIKNFGEDMDDE--RLKD 209
> dre:393856 pabpc1b, MGC77608, pabpc1, zgc:77608; poly A binding
protein, cytoplasmic 1 b; K13126 polyadenylate-binding protein
Length=634
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 29/116 (25%), Positives = 54/116 (46%), Gaps = 4/116 (3%)
Query 1 QQQQQQQQQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFC 60
+Q +Q + + + LY+ N+ D++ +R FG + D G+ FGF
Sbjct 281 EQMKQDRMTRYQGVNLYVKNLDDGIDDERLRKEFSPFGTITSAKVMMD--GGRSKGFGFV 338
Query 61 DFAEPLAAAKCVACLAGRKLHGRSLVVNCNEKVRAKIAKVTEERI--LATMRAYKN 114
F+ P A K V + GR + + L V ++ + A +T + + +A++RA N
Sbjct 339 CFSSPEEATKAVTEMNGRIVATKPLYVALAQRKEERQAHLTNQYMQRMASVRAVPN 394
Score = 36.6 bits (83), Expect = 0.029, Method: Compositional matrix adjust.
Identities = 22/89 (24%), Positives = 46/89 (51%), Gaps = 4/89 (4%)
Query 14 TTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVA 73
T +YI N + D+D ++++ ++G + +D +G+ FGF F A + V
Sbjct 191 TNVYIKNFGEDMDDDKLKDIFSKYGNAMSIRVMTDE-NGKSRGFGFVSFERHEDAQRAVD 249
Query 74 CLAGRKLHGRSLVVNCNEKVRAKIAKVTE 102
+ G++++G+ + V +K K+ + TE
Sbjct 250 EMNGKEMNGKLIYVGRAQK---KVERQTE 275
Score = 28.9 bits (63), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 18/72 (25%), Positives = 31/72 (43%), Gaps = 2/72 (2%)
Query 16 LYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVACL 75
++I N+ K DN + + FG +L D + +GF F AA + + +
Sbjct 101 IFIKNLDKSIDNKALYDTFSAFGNILSCKVVCDENGSK--GYGFVHFETQEAAERAIEKM 158
Query 76 AGRKLHGRSLVV 87
G L+ R + V
Sbjct 159 NGMLLNDRKVFV 170
> tpv:TP02_0374 spliceosome associated protein; K12831 splicing
factor 3B subunit 4
Length=290
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 24/74 (32%), Positives = 37/74 (50%), Gaps = 0/74 (0%)
Query 15 TLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVAC 74
TLYIGN+ AD + + + GRV N D ++GQ FGF ++ + A +
Sbjct 19 TLYIGNLDLQADEELLWEFFMQAGRVKSINVPRDKVTGQHQGFGFVEYETEVDADYALKI 78
Query 75 LAGRKLHGRSLVVN 88
L KL+ + L +N
Sbjct 79 LNFVKLYHKPLKLN 92
> sce:YER165W PAB1; Pab1p; K13126 polyadenylate-binding protein
Length=577
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 30/129 (23%), Positives = 59/129 (45%), Gaps = 1/129 (0%)
Query 13 ATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCV 72
+ ++I N+ DN + + FG +L +D +G+ FGF F E AA + +
Sbjct 125 SGNIFIKNLHPDIDNKALYDTFSVFGDILSSKIATDE-NGKSKGFGFVHFEEEGAAKEAI 183
Query 73 ACLAGRKLHGRSLVVNCNEKVRAKIAKVTEERILATMRAYKNKTREQVEKEIEEEYKRLK 132
L G L+G+ + V + + + +++ E + T KN E +++ +E + +
Sbjct 184 DALNGMLLNGQEIYVAPHLSRKERDSQLEETKAHYTNLYVKNINSETTDEQFQELFAKFG 243
Query 133 DGVQQILDK 141
V L+K
Sbjct 244 PIVSASLEK 252
Score = 42.4 bits (98), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 23/83 (27%), Positives = 41/83 (49%), Gaps = 1/83 (1%)
Query 5 QQQQQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAE 64
Q ++ + T LY+ NI ++ + L +FG ++ + + D G+L FGF ++ +
Sbjct 210 QLEETKAHYTNLYVKNINSETTDEQFQELFAKFGPIVSASLEKD-ADGKLKGFGFVNYEK 268
Query 65 PLAAAKCVACLAGRKLHGRSLVV 87
A K V L +L+G L V
Sbjct 269 HEDAVKAVEALNDSELNGEKLYV 291
> hsa:80336 PABPC1L, C20orf119, FLJ11840, FLJ30809, FLJ42053,
PABPC1L1, dJ1069P2.3, ePAB; poly(A) binding protein, cytoplasmic
1-like; K13126 polyadenylate-binding protein
Length=614
Score = 43.1 bits (100), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 29/116 (25%), Positives = 54/116 (46%), Gaps = 4/116 (3%)
Query 1 QQQQQQQQQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFC 60
+Q +Q + ++ + LY+ N+ D+D +R +G + ++ G FGF
Sbjct 281 EQMKQDRLRRYQGVNLYVKNLDDSIDDDKLRKEFSPYGVITSAKVMTE--GGHSKGFGFV 338
Query 61 DFAEPLAAAKCVACLAGRKLHGRSLVVNCNEKVRAKIAKVTEERI--LATMRAYKN 114
F+ P A K V + GR + + L V ++ + A +T + + L+TMR N
Sbjct 339 CFSSPEEATKAVTEMNGRIVGTKPLYVALAQRKEERKAILTNQYMQRLSTMRTLSN 394
Score = 38.9 bits (89), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 22/72 (30%), Positives = 37/72 (51%), Gaps = 1/72 (1%)
Query 14 TTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVA 73
T +Y+ N+ D +++L +FG++L D SG FGF +F + A K V
Sbjct 191 TNIYVKNLPVDVDEQGLQDLFSQFGKMLSVKVMRD-NSGHSRCFGFVNFEKHEEAQKAVV 249
Query 74 CLAGRKLHGRSL 85
+ G+++ GR L
Sbjct 250 HMNGKEVSGRLL 261
> ath:AT5G50250 31 kDa ribonucleoprotein, chloroplast, putative
/ RNA-binding protein RNP-T, putative / RNA-binding protein
1/2/3, putative / RNA-binding protein cp31, putative
Length=289
Score = 43.1 bits (100), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 25/92 (27%), Positives = 44/92 (47%), Gaps = 0/92 (0%)
Query 1 QQQQQQQQQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFC 60
+ ++Q + A +Y+GN+ D+ + L E G+V+ SD +G+ FGF
Sbjct 194 SRPERQPRVYDAAFRIYVGNLPWDVDSGRLERLFSEHGKVVDARVVSDRETGRSRGFGFV 253
Query 61 DFAEPLAAAKCVACLAGRKLHGRSLVVNCNEK 92
+ +A L G+ L GR++ VN E+
Sbjct 254 QMSNENEVNVAIAALDGQNLEGRAIKVNVAEE 285
Score = 30.8 bits (68), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 22/77 (28%), Positives = 35/77 (45%), Gaps = 8/77 (10%)
Query 16 LYIGNITKHADNDFMRNLLGEFGRV----LRWNRQSDPISGQLAAFGFCDFAEPLAAAKC 71
L++GN+ D+ + L + G V + +NR +D Q FGF + A K
Sbjct 115 LFVGNLPYDVDSQALAMLFEQAGTVEISEVIYNRDTD----QSRGFGFVTMSTVEEAEKA 170
Query 72 VACLAGRKLHGRSLVVN 88
V +++GR L VN
Sbjct 171 VEKFNSFEVNGRRLTVN 187
> ath:AT4G24770 RBP31; RBP31 (31-KDA RNA BINDING PROTEIN); RNA
binding / poly(U) binding
Length=329
Score = 43.1 bits (100), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 22/77 (28%), Positives = 39/77 (50%), Gaps = 0/77 (0%)
Query 16 LYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVACL 75
+Y+GN+ DN + L E G+V+ D +G+ FGF ++ + ++ L
Sbjct 246 VYVGNLPWDVDNGRLEQLFSEHGKVVEARVVYDRETGRSRGFGFVTMSDVDELNEAISAL 305
Query 76 AGRKLHGRSLVVNCNEK 92
G+ L GR++ VN E+
Sbjct 306 DGQNLEGRAIRVNVAEE 322
> cel:Y47G6A.20 rnp-6; RNP (RRM RNA binding domain) containing
family member (rnp-6); K12838 poly(U)-binding-splicing factor
PUF60
Length=749
Score = 43.1 bits (100), Expect = 3e-04, Method: Composition-based stats.
Identities = 23/75 (30%), Positives = 36/75 (48%), Gaps = 0/75 (0%)
Query 14 TTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVA 73
+ +Y+G+I+ D +R FG + N DP +G F F ++ P AA
Sbjct 102 SRIYVGSISFEIREDMLRRAFDPFGPIKSINMSWDPATGHHKTFAFVEYEVPEAALLAQE 161
Query 74 CLAGRKLHGRSLVVN 88
+ G+ L GR+L VN
Sbjct 162 SMNGQMLGGRNLKVN 176
> mmu:18458 Pabpc1, PABP, Pabp1, PabpI, Pabpl1, ePAB; poly(A)
binding protein, cytoplasmic 1; K13126 polyadenylate-binding
protein
Length=636
Score = 43.1 bits (100), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 25/79 (31%), Positives = 41/79 (51%), Gaps = 1/79 (1%)
Query 14 TTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVA 73
T +YI N + D++ ++ L G+FG L +D SG+ FGF F A K V
Sbjct 191 TNVYIKNFGEDMDDERLKELFGKFGPALSVKVMTDE-SGKSKGFGFVSFERHEDAQKAVD 249
Query 74 CLAGRKLHGRSLVVNCNEK 92
+ G++L+G+ + V +K
Sbjct 250 EMNGKELNGKQIYVGRAQK 268
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 28/116 (24%), Positives = 54/116 (46%), Gaps = 4/116 (3%)
Query 1 QQQQQQQQQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFC 60
+Q +Q + + + LY+ N+ D++ +R FG + + G+ FGF
Sbjct 281 EQMKQDRITRYQGVNLYVKNLDDGIDDERLRKEFSPFGTITSAKVMME--GGRSKGFGFV 338
Query 61 DFAEPLAAAKCVACLAGRKLHGRSLVVNCNEKVRAKIAKVTEERI--LATMRAYKN 114
F+ P A K V + GR + + L V ++ + A +T + + +A++RA N
Sbjct 339 CFSSPEEATKAVTEMNGRIVATKPLYVALAQRKEERQAHLTNQYMQRMASVRAVPN 394
Score = 28.5 bits (62), Expect = 8.4, Method: Compositional matrix adjust.
Identities = 28/118 (23%), Positives = 51/118 (43%), Gaps = 9/118 (7%)
Query 16 LYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVACL 75
++I N+ K DN + + FG +L D + +GF F AA + + +
Sbjct 101 IFIKNLDKSIDNKALYDTFSAFGNILSCKVVCDENGSK--GYGFVHFETQEAAERAIEKM 158
Query 76 AGRKLHGRSLVVNCNEKVRAKIAKVTEERILATMRAYKNKTREQVEKEIEEEYKRLKD 133
G L+ R + V R K K E + A + + N + +++++E RLK+
Sbjct 159 NGMLLNDRKVFVG-----RFKSRKEREAELGARAKEFTNVYIKNFGEDMDDE--RLKE 209
> hsa:5042 PABPC3, PABP3, PABPL3, tPABP; poly(A) binding protein,
cytoplasmic 3; K13126 polyadenylate-binding protein
Length=631
Score = 43.1 bits (100), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 37/121 (30%), Positives = 61/121 (50%), Gaps = 9/121 (7%)
Query 16 LYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVACL 75
+YI N + D++ +++L G+FG L +D SG+ FGF F A K V +
Sbjct 193 VYIKNFGEDMDDERLKDLFGKFGPALSVKVMTDE-SGKSKGFGFVSFERHEDAQKAVDEM 251
Query 76 AGRKLHGRSLVVNCNEKVRAKIAKVTE-ERILATMRAYKNKTREQVEKEIEEEYKRLKDG 134
G++L+G+ + V +K K+ + TE +R M+ + TR QV + K L DG
Sbjct 252 NGKELNGKQIYVGRAQK---KVERQTELKRTFEQMKQDR-ITRYQV---VNLYVKNLDDG 304
Query 135 V 135
+
Sbjct 305 I 305
Score = 42.7 bits (99), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 29/117 (24%), Positives = 55/117 (47%), Gaps = 4/117 (3%)
Query 1 QQQQQQQQQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFC 60
+Q +Q + + + LY+ N+ D++ +R FG + + G+ FGF
Sbjct 281 EQMKQDRITRYQVVNLYVKNLDDGIDDERLRKAFSPFGTITSAKVMME--GGRSKGFGFV 338
Query 61 DFAEPLAAAKCVACLAGRKLHGRSLVVNCNEKVRAKIAKVTEERI--LATMRAYKNK 115
F+ P A K V + GR + + L V ++ + A +T E + +A++RA N+
Sbjct 339 CFSSPEEATKAVTEMNGRIVATKPLYVALAQRKEERQAYLTNEYMQRMASVRAVPNQ 395
Score = 32.3 bits (72), Expect = 0.64, Method: Compositional matrix adjust.
Identities = 28/118 (23%), Positives = 54/118 (45%), Gaps = 9/118 (7%)
Query 16 LYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVACL 75
+++ N+ K +N + + + FG +L N D + +GF F AA + + +
Sbjct 101 IFVKNLDKSINNKALYDTVSAFGNILSCNVVCDENGSK--GYGFVHFETHEAAERAIKKM 158
Query 76 AGRKLHGRSLVVNCNEKVRAKIAKVTEERILATMRAYKNKTREQVEKEIEEEYKRLKD 133
G L+GR + V + K K E + A + + N + +++++E RLKD
Sbjct 159 NGMLLNGRKVFVG-----QFKSRKEREAELGARAKEFPNVYIKNFGEDMDDE--RLKD 209
> xla:495336 hypothetical LOC495336
Length=711
Score = 43.1 bits (100), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 22/82 (26%), Positives = 41/82 (50%), Gaps = 1/82 (1%)
Query 4 QQQQQQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFA 63
++ Q ++Q+ +Y+ N D++ ++ + EFG + D G+ FGF +
Sbjct 171 RKSQNRKQKFNNIYVKNFPPETDDEKLKEMFTEFGEIKSACVMKD-SEGKSKGFGFVCYL 229
Query 64 EPLAAAKCVACLAGRKLHGRSL 85
P A VA + G+++ GRSL
Sbjct 230 NPEHAEAAVAAMHGKEIGGRSL 251
Score = 32.3 bits (72), Expect = 0.61, Method: Compositional matrix adjust.
Identities = 17/74 (22%), Positives = 36/74 (48%), Gaps = 0/74 (0%)
Query 14 TTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVA 73
T+LY+G++ ++D +R E G V + D S + +G+ +F +P A + +
Sbjct 2 TSLYVGDLHPDINDDQLRMKFSEIGPVAVAHVCRDVTSRKSLGYGYVNFEDPKDAERALE 61
Query 74 CLAGRKLHGRSLVV 87
+ + GR + +
Sbjct 62 QMNYEVVMGRPIRI 75
> hsa:23283 CSTF2T, CstF-64T, DKFZp434C1013, KIAA0689; cleavage
stimulation factor, 3' pre-RNA, subunit 2, 64kDa, tau variant;
K14407 cleavage stimulation factor subunit 2
Length=616
Score = 43.1 bits (100), Expect = 4e-04, Method: Composition-based stats.
Identities = 20/74 (27%), Positives = 41/74 (55%), Gaps = 0/74 (0%)
Query 15 TLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVAC 74
++++GNI A + ++++ E G V+ + D +G+ +GFC++ + A +
Sbjct 17 SVFVGNIPYEATEEQLKDIFSEVGSVVSFRLVYDRETGKPKGYGFCEYQDQETALSAMRN 76
Query 75 LAGRKLHGRSLVVN 88
L GR+ GR+L V+
Sbjct 77 LNGREFSGRALRVD 90
> mmu:83410 Cstf2t, 64kDa, C77975, tCstF-64, tauCstF-64; cleavage
stimulation factor, 3' pre-RNA subunit 2, tau; K14407 cleavage
stimulation factor subunit 2
Length=632
Score = 42.7 bits (99), Expect = 4e-04, Method: Composition-based stats.
Identities = 20/74 (27%), Positives = 41/74 (55%), Gaps = 0/74 (0%)
Query 15 TLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVAC 74
++++GNI A + ++++ E G V+ + D +G+ +GFC++ + A +
Sbjct 17 SVFVGNIPYEATEEQLKDIFSEVGSVVSFRLVYDRETGKPKGYGFCEYQDQETALSAMRN 76
Query 75 LAGRKLHGRSLVVN 88
L GR+ GR+L V+
Sbjct 77 LNGREFSGRALRVD 90
> sce:YNL016W PUB1, RNP1; Poly (A)+ RNA-binding protein, abundant
mRNP-component protein that binds mRNA and is required for
stability of many mRNAs; component of glucose deprivation
induced stress granules, involved in P-body-dependent granule
assembly; K13201 nucleolysin TIA-1/TIAR
Length=453
Score = 42.7 bits (99), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 23/89 (25%), Positives = 40/89 (44%), Gaps = 0/89 (0%)
Query 4 QQQQQQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFA 63
Q QQ L++G++ + D++ +RN +F L + D +G +GF F
Sbjct 152 QSQQSSSDDTFNLFVGDLNVNVDDETLRNAFKDFPSYLSGHVMWDMQTGSSRGYGFVSFT 211
Query 64 EPLAAAKCVACLAGRKLHGRSLVVNCNEK 92
A + + G+ L+GR L +N K
Sbjct 212 SQDDAQNAMDSMQGQDLNGRPLRINWAAK 240
Score = 31.6 bits (70), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 22/74 (29%), Positives = 29/74 (39%), Gaps = 6/74 (8%)
Query 12 RATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKC 71
R TT YIGNI A + L FG +L + + F + AA C
Sbjct 339 RVTTAYIGNIPHFATEADLIPLFQNFGFILDFKHYPE------KGCCFIKYDTHEQAAVC 392
Query 72 VACLAGRKLHGRSL 85
+ LA GR+L
Sbjct 393 IVALANFPFQGRNL 406
> hsa:8761 PABPC4, APP-1, APP1, FLJ43938, PABP4, iPABP; poly(A)
binding protein, cytoplasmic 4 (inducible form); K13126 polyadenylate-binding
protein
Length=660
Score = 42.7 bits (99), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 26/99 (26%), Positives = 50/99 (50%), Gaps = 8/99 (8%)
Query 14 TTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVA 73
T +YI N + D++ ++ L +FG+ L DP +G+ FGF + + A K V
Sbjct 191 TNVYIKNFGEEVDDESLKELFSQFGKTLSVKVMRDP-NGKSKGFGFVSYEKHEDANKAVE 249
Query 74 CLAGRKLHGRSLVVNCNEK-------VRAKIAKVTEERI 105
+ G+++ G+ + V +K ++ K ++ +ERI
Sbjct 250 EMNGKEISGKIIFVGRAQKKVERQAELKRKFEQLKQERI 288
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 29/113 (25%), Positives = 53/113 (46%), Gaps = 4/113 (3%)
Query 1 QQQQQQQQQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFC 60
+Q +Q++ + + LYI N+ D++ +R FG + + G+ FGF
Sbjct 281 EQLKQERISRYQGVNLYIKNLDDTIDDEKLRKEFSPFGSITSAKVMLE--DGRSKGFGFV 338
Query 61 DFAEPLAAAKCVACLAGRKLHGRSLVVNCNEKVRAKIAKVTEERI--LATMRA 111
F+ P A K V + GR + + L V ++ + A +T + + +A MRA
Sbjct 339 CFSSPEEATKAVTEMNGRIVGSKPLYVALAQRKEERKAHLTNQYMQRVAGMRA 391
> ath:AT4G34110 PAB2; PAB2 (POLY(A) BINDING 2); RNA binding /
translation initiation factor; K13126 polyadenylate-binding
protein
Length=629
Score = 42.4 bits (98), Expect = 6e-04, Method: Composition-based stats.
Identities = 29/122 (23%), Positives = 55/122 (45%), Gaps = 9/122 (7%)
Query 1 QQQQQQQQQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFC 60
+Q++ + + T +Y+ N+ + +D ++N GE+G++ D G+ FGF
Sbjct 202 RQERDSTANKTKFTNVYVKNLAESTTDDDLKNAFGEYGKITSAVVMKDG-EGKSKGFGFV 260
Query 61 DFAEPLAAAKCVACLAGRKLHGRSLVVNCNEKVRAKIAKVTEERILATMRAYKNKTREQV 120
+F AA+ V L G K + V + + K + TE R+ Y+ +E
Sbjct 261 NFENADDAARAVESLNGHKFDDKEWYVG---RAQKKSERETELRV-----RYEQNLKEAA 312
Query 121 EK 122
+K
Sbjct 313 DK 314
Score = 35.8 bits (81), Expect = 0.049, Method: Composition-based stats.
Identities = 20/87 (22%), Positives = 42/87 (48%), Gaps = 1/87 (1%)
Query 1 QQQQQQQQQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFC 60
+Q ++ + +++ LY+ N+ ++ ++ + FG V DP +G GF
Sbjct 305 EQNLKEAADKFQSSNLYVKNLDPSISDEKLKEIFSPFGTVTSSKVMRDP-NGTSKGSGFV 363
Query 61 DFAEPLAAAKCVACLAGRKLHGRSLVV 87
FA P A + ++ L+G+ + + L V
Sbjct 364 AFATPEEATEAMSQLSGKMIESKPLYV 390
Score = 31.2 bits (69), Expect = 1.3, Method: Composition-based stats.
Identities = 17/74 (22%), Positives = 38/74 (51%), Gaps = 0/74 (0%)
Query 14 TTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVA 73
T+LY+G++ + + + + G+ G V+ D ++ + +G+ +F P AA+ +
Sbjct 36 TSLYVGDLDFNVTDSQLFDAFGQMGTVVTVRVCRDLVTRRSLGYGYVNFTNPQDAARAIQ 95
Query 74 CLAGRKLHGRSLVV 87
L L+G+ + V
Sbjct 96 ELNYIPLYGKPIRV 109
Score = 31.2 bits (69), Expect = 1.5, Method: Composition-based stats.
Identities = 19/75 (25%), Positives = 35/75 (46%), Gaps = 1/75 (1%)
Query 13 ATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCV 72
A ++I N+ + D+ + + FG ++ D SGQ +GF +A +A K +
Sbjct 123 AGNIFIKNLDESIDHKALHDTFSSFGNIVSCKVAVDS-SGQSKGYGFVQYANEESAQKAI 181
Query 73 ACLAGRKLHGRSLVV 87
L G L+ + + V
Sbjct 182 EKLNGMLLNDKQVYV 196
> mmu:67543 Pabpc6, 4932702K14Rik, AI428050, MGC132900, Pabpc3;
poly(A) binding protein, cytoplasmic 6; K13126 polyadenylate-binding
protein
Length=643
Score = 42.4 bits (98), Expect = 6e-04, Method: Composition-based stats.
Identities = 28/101 (27%), Positives = 51/101 (50%), Gaps = 4/101 (3%)
Query 2 QQQQQQQQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCD 61
+Q + + + T +YI N+ + D++ +++L G FG L +D SG+ FGF
Sbjct 179 RQAELGARAKEFTNVYIKNLGEDMDDERLQDLFGRFGPALSVKVMTDE-SGKSKGFGFVS 237
Query 62 FAEPLAAAKCVACLAGRKLHGRSLVVNCNEKVRAKIAKVTE 102
F A K V + G+ L+G+ + V + + K+ + TE
Sbjct 238 FERHEDARKAVEEMNGKDLNGKQIYVG---RAQKKVERQTE 275
Score = 31.2 bits (69), Expect = 1.4, Method: Composition-based stats.
Identities = 21/99 (21%), Positives = 43/99 (43%), Gaps = 2/99 (2%)
Query 5 QQQQQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAE 64
Q + + + LY+ N+ D++ +R FG + + G+ FGF F+
Sbjct 295 QDRSVRCKGVNLYVKNLDDGIDDERLRKEFSPFGTITSAKVTME--GGRSKGFGFVCFSS 352
Query 65 PLAAAKCVACLAGRKLHGRSLVVNCNEKVRAKIAKVTEE 103
P A K V + G+ + + L V ++ + A ++ +
Sbjct 353 PEEATKAVTEMNGKIVATKPLYVALAQRKEERQAHLSNQ 391
> mmu:18459 Pabpc2, PABP, PABP+, Pabp2; poly(A) binding protein,
cytoplasmic 2; K13126 polyadenylate-binding protein
Length=628
Score = 42.4 bits (98), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 27/100 (27%), Positives = 49/100 (49%), Gaps = 8/100 (8%)
Query 14 TTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVA 73
T +YI N D++ + L G FG++L +D G+ FGF F A K V
Sbjct 191 TNVYIKNFGDRMDDETLNGLFGRFGQILSVKVMTDE-GGKSKGFGFVSFERHEDAQKAVD 249
Query 74 CLAGRKLHGRSLVVNCNEK-------VRAKIAKVTEERIL 106
+ G++L+G+ + V +K ++ K +VT+++ +
Sbjct 250 EMNGKELNGKHIYVGRAQKKDDRHTELKHKFEQVTQDKSI 289
Score = 38.9 bits (89), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 27/116 (23%), Positives = 55/116 (47%), Gaps = 4/116 (3%)
Query 1 QQQQQQQQQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFC 60
+Q Q + + + LY+ N+ D++ ++ FG + ++ G+ FGF
Sbjct 281 EQVTQDKSIRYQGINLYVKNLDDGIDDERLQKEFSPFGTITSTKVMTE--GGRSKGFGFV 338
Query 61 DFAEPLAAAKCVACLAGRKLHGRSLVVNCNEKVRAKIAKVTEERI--LATMRAYKN 114
F+ P A K V+ + GR + + L V ++ + A +T + I +A++R+ N
Sbjct 339 CFSSPEEATKAVSEMNGRIVATKPLYVALAQRKEERQAHLTNQYIQRMASVRSGPN 394
Score = 28.5 bits (62), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 19/72 (26%), Positives = 31/72 (43%), Gaps = 2/72 (2%)
Query 16 LYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVACL 75
++I N+ K DN + + FG +L SD + GF F AA + + +
Sbjct 101 VFIKNLNKTIDNKALYDTFSAFGNILSCKVVSDENGSK--GHGFVHFETEEAAERAIEKM 158
Query 76 AGRKLHGRSLVV 87
G L+ R + V
Sbjct 159 NGMLLNDRKVFV 170
> xla:379871 cstf2, MGC53575, MGC83019, cstf-64; cleavage stimulation
factor, 3' pre-RNA, subunit 2, 64kDa; K14407 cleavage
stimulation factor subunit 2
Length=518
Score = 42.4 bits (98), Expect = 6e-04, Method: Composition-based stats.
Identities = 20/74 (27%), Positives = 41/74 (55%), Gaps = 0/74 (0%)
Query 15 TLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVAC 74
++++GNI A + ++++ E G V+ + D +G+ +GFC++ + A +
Sbjct 17 SVFVGNIPYEATEEQLKDIFSEVGPVVSFRLVYDRETGKPKGYGFCEYQDQETALSAMRN 76
Query 75 LAGRKLHGRSLVVN 88
L GR+ GR+L V+
Sbjct 77 LNGREFSGRALRVD 90
> ath:AT4G13850 GR-RBP2 (GLYCINE-RICH RNA-BINDING PROTEIN 2);
ATP binding / RNA binding / double-stranded DNA binding / single-stranded
DNA binding
Length=144
Score = 42.4 bits (98), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 23/76 (30%), Positives = 42/76 (55%), Gaps = 0/76 (0%)
Query 13 ATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCV 72
+T L+IG ++ D+ +R+ FG V+ D +G+ FGF +F + AA +
Sbjct 34 STKLFIGGLSWGTDDASLRDAFAHFGDVVDAKVIVDRETGRSRGFGFVNFNDEGAATAAI 93
Query 73 ACLAGRKLHGRSLVVN 88
+ + G++L+GR + VN
Sbjct 94 SEMDGKELNGRHIRVN 109
> dre:386806 cstf2, CstF-64, fb11e07, wu:fb11e07, zgc:56346, zgc:77730;
cleavage stimulation factor, 3' pre-RNA, subunit 2;
K14407 cleavage stimulation factor subunit 2
Length=488
Score = 42.4 bits (98), Expect = 7e-04, Method: Composition-based stats.
Identities = 20/74 (27%), Positives = 41/74 (55%), Gaps = 0/74 (0%)
Query 15 TLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVAC 74
++++GNI A + ++++ E G V+ + D +G+ +GFC++ + A +
Sbjct 25 SVFVGNIPYEATEEQLKDIFSEVGLVVSFRLVYDRETGKPKGYGFCEYQDQETALSAMRN 84
Query 75 LAGRKLHGRSLVVN 88
L GR+ GR+L V+
Sbjct 85 LNGREFSGRALRVD 98
> xla:398221 pabpc1l-a, MGC82273, ePAB, epabp-a, pabpc1l1; poly(A)
binding protein, cytoplasmic 1-like; K13126 polyadenylate-binding
protein
Length=629
Score = 42.4 bits (98), Expect = 7e-04, Method: Composition-based stats.
Identities = 30/113 (26%), Positives = 54/113 (47%), Gaps = 4/113 (3%)
Query 1 QQQQQQQQQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFC 60
+Q +Q++ + + LY+ N+ D+D +R +G + ++ G FGF
Sbjct 281 EQIKQERINRYQGVNLYVKNLDDGIDDDRLRKEFLPYGTITSAKVMTE--GGHSKGFGFV 338
Query 61 DFAEPLAAAKCVACLAGRKLHGRSLVVNCNEKVRAKIAKVTEERI--LATMRA 111
F+ P A K V + GR + + L V ++ + A +T + + LATMRA
Sbjct 339 CFSSPEEATKAVTEMNGRIVSTKPLYVALAQRKEERKAILTNQYMQRLATMRA 391
Score = 40.0 bits (92), Expect = 0.003, Method: Composition-based stats.
Identities = 27/99 (27%), Positives = 48/99 (48%), Gaps = 8/99 (8%)
Query 14 TTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVA 73
T +YI N + D+ +R + FG L D SG+ FGF ++ A K V+
Sbjct 191 TNVYIKNFGEDMDDKRLREIFSAFGNTLSVKVMMDD-SGRSRGFGFVNYGNHEEAQKAVS 249
Query 74 CLAGRKLHGRSLVVNCNEK-------VRAKIAKVTEERI 105
+ G++++GR + V +K ++ K ++ +ERI
Sbjct 250 EMNGKEVNGRMIYVGRAQKRIERQSELKRKFEQIKQERI 288
> dre:606498 pabpc1a, pabpc1, wu:fb16a02, wu:fi19b08, wu:fj12d09,
wu:fj61f06, zgc:109879; poly A binding protein, cytoplasmic
1 a; K13126 polyadenylate-binding protein
Length=634
Score = 42.0 bits (97), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 28/118 (23%), Positives = 54/118 (45%), Gaps = 4/118 (3%)
Query 1 QQQQQQQQQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFC 60
+Q +Q + + + LY+ N+ D++ +R FG + + G+ FGF
Sbjct 281 EQMKQDRMTRYQGVNLYVKNLDDGLDDERLRKEFSPFGTITSAKVMME--GGRSKGFGFV 338
Query 61 DFAEPLAAAKCVACLAGRKLHGRSLVVNCNEKVRAKIAKVTEERI--LATMRAYKNKT 116
F+ P A K V + GR + + L V ++ + A +T + + +A++RA N
Sbjct 339 CFSSPEEATKAVTEMNGRIVATKPLYVALAQRKEERQAHLTSQYMQRMASVRAVPNPV 396
Score = 35.0 bits (79), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 20/79 (25%), Positives = 40/79 (50%), Gaps = 1/79 (1%)
Query 14 TTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVA 73
T +YI N + D++ ++ + ++G L +D SG+ FGF F A + V
Sbjct 191 TNVYIKNFGEDMDDEKLKEIFCKYGPALSIRVMTD-DSGKSKGFGFVSFERHEDAQRAVD 249
Query 74 CLAGRKLHGRSLVVNCNEK 92
+ G++++G+ + V +K
Sbjct 250 EMNGKEMNGKQVYVGRAQK 268
Score = 28.5 bits (62), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 18/72 (25%), Positives = 31/72 (43%), Gaps = 2/72 (2%)
Query 16 LYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVACL 75
++I N+ K DN + + FG +L D + +GF F AA + + +
Sbjct 101 IFIKNLDKSIDNKALYDTFSAFGNILSCKVVCDENGSK--GYGFVHFETHEAAERAIEKM 158
Query 76 AGRKLHGRSLVV 87
G L+ R + V
Sbjct 159 NGMLLNDRKVFV 170
> hsa:1478 CSTF2, CstF-64; cleavage stimulation factor, 3' pre-RNA,
subunit 2, 64kDa; K14407 cleavage stimulation factor subunit
2
Length=577
Score = 42.0 bits (97), Expect = 7e-04, Method: Composition-based stats.
Identities = 20/74 (27%), Positives = 41/74 (55%), Gaps = 0/74 (0%)
Query 15 TLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVAC 74
++++GNI A + ++++ E G V+ + D +G+ +GFC++ + A +
Sbjct 17 SVFVGNIPYEATEEQLKDIFSEVGPVVSFRLVYDRETGKPKGYGFCEYQDQETALSAMRN 76
Query 75 LAGRKLHGRSLVVN 88
L GR+ GR+L V+
Sbjct 77 LNGREFSGRALRVD 90
> ath:AT5G54580 RNA recognition motif (RRM)-containing protein
Length=156
Score = 42.0 bits (97), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 22/82 (26%), Positives = 41/82 (50%), Gaps = 0/82 (0%)
Query 1 QQQQQQQQQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFC 60
+ Q + Q + +T L++ ++K ++ +R +FG V +D +SG FGF
Sbjct 43 ESQTPARPQAEPSTNLFVSGLSKRTTSEGLRTAFAQFGEVADAKVVTDRVSGYSKGFGFV 102
Query 61 DFAEPLAAAKCVACLAGRKLHG 82
+A +AK +A + G+ L G
Sbjct 103 RYATLEDSAKGIAGMDGKFLDG 124
> tgo:TGME49_068750 peptidyl-prolyl cis-trans isomerase E, putative
Length=145
Score = 42.0 bits (97), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 21/76 (27%), Positives = 38/76 (50%), Gaps = 0/76 (0%)
Query 15 TLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVAC 74
TLY+G + + + + +R FG + + D +G FGF +F E A + +
Sbjct 22 TLYVGGLAEQVEEEVLRAAFLPFGDIKQLEIPKDKTTGLHRGFGFVEFEEEDDAKEAMEN 81
Query 75 LAGRKLHGRSLVVNCN 90
+ +L+GR+L VN +
Sbjct 82 MDNAELYGRTLRVNLS 97
> ath:AT5G51300 splicing factor-related; K13095 splicing factor
1
Length=804
Score = 42.0 bits (97), Expect = 9e-04, Method: Composition-based stats.
Identities = 22/74 (29%), Positives = 36/74 (48%), Gaps = 0/74 (0%)
Query 14 TTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVA 73
T LYIG + ++D + NL FG ++ D ++G +GF +A+ A V
Sbjct 480 TNLYIGFLPPMLEDDGLINLFSSFGEIVMAKVIKDRVTGLSKGYGFVKYADVQMANTAVQ 539
Query 74 CLAGRKLHGRSLVV 87
+ G + GR+L V
Sbjct 540 AMNGYRFEGRTLAV 553
> xla:380287 u2af2, MGC53441, u2af65; U2 small nuclear RNA auxiliary
factor 2; K12837 splicing factor U2AF 65 kDa subunit
Length=456
Score = 42.0 bits (97), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 20/75 (26%), Positives = 39/75 (52%), Gaps = 0/75 (0%)
Query 13 ATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCV 72
A L+IG + + ++D ++ LL FG + +N D +G + FC++ + + +
Sbjct 243 AHKLFIGGLPNYLNDDQVKELLTSFGPLKAFNLVKDSATGLSKGYAFCEYVDINVTDQAI 302
Query 73 ACLAGRKLHGRSLVV 87
A L G +L + L+V
Sbjct 303 AGLNGMQLGDKKLLV 317
> mmu:108062 Cstf2, 64kDa, C630034J23Rik, Cstf64; cleavage stimulation
factor, 3' pre-RNA subunit 2; K14407 cleavage stimulation
factor subunit 2
Length=580
Score = 42.0 bits (97), Expect = 9e-04, Method: Composition-based stats.
Identities = 20/74 (27%), Positives = 41/74 (55%), Gaps = 0/74 (0%)
Query 15 TLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVAC 74
++++GNI A + ++++ E G V+ + D +G+ +GFC++ + A +
Sbjct 17 SVFVGNIPYEATEEQLKDIFSEVGPVVSFRLVYDRETGKPKGYGFCEYQDQETALSAMRN 76
Query 75 LAGRKLHGRSLVVN 88
L GR+ GR+L V+
Sbjct 77 LNGREFSGRALRVD 90
> hsa:132430 PABPC4L, DKFZp686J06116; poly(A) binding protein,
cytoplasmic 4-like; K13126 polyadenylate-binding protein
Length=428
Score = 41.6 bits (96), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 22/74 (29%), Positives = 39/74 (52%), Gaps = 1/74 (1%)
Query 14 TTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVA 73
T +YI N D++ ++++ ++G+ L +D SG+ FGF F AA K V
Sbjct 248 TNVYIKNFGGDMDDERLKDVFSKYGKTLSVKVMTDS-SGKSKGFGFVSFDSHEAAKKAVE 306
Query 74 CLAGRKLHGRSLVV 87
+ GR ++G+ + V
Sbjct 307 EMNGRDINGQLIFV 320
Score = 36.2 bits (82), Expect = 0.042, Method: Compositional matrix adjust.
Identities = 22/75 (29%), Positives = 33/75 (44%), Gaps = 2/75 (2%)
Query 13 ATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCV 72
LYI N+ D++ +RN FG + R + GQ FG F+ P A K +
Sbjct 350 GVKLYIKNLDDTIDDEKLRNEFSSFGSISRVKVMQEE--GQSKGFGLICFSSPEDATKAM 407
Query 73 ACLAGRKLHGRSLVV 87
+ GR L + L +
Sbjct 408 TEMNGRILGSKPLSI 422
> cel:F56A8.6 cpf-2; Cleavage and Polyadenylation Factor family
member (cpf-2); K14407 cleavage stimulation factor subunit
2
Length=336
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 21/75 (28%), Positives = 38/75 (50%), Gaps = 0/75 (0%)
Query 15 TLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVAC 74
++++GNI+ D +R++ + G VL D +G+ +GF +F + A +
Sbjct 19 SVFVGNISYDVSEDTIRSIFSKAGNVLSIKMVHDRETGKPKGYGFIEFPDIQTAEVAIRN 78
Query 75 LAGRKLHGRSLVVNC 89
L G +L GR L V+
Sbjct 79 LNGYELSGRILRVDS 93
> xla:444284 pabpc4, MGC80927, PABP, ePAB, ePABP; poly(A) binding
protein, cytoplasmic 4 (inducible form); K13126 polyadenylate-binding
protein
Length=626
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 30/118 (25%), Positives = 54/118 (45%), Gaps = 4/118 (3%)
Query 1 QQQQQQQQQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFC 60
+Q +Q++ + + LYI N+ D++ +R FG + + G+ FGF
Sbjct 281 EQLKQERISRYQGVNLYIKNLDDTIDDEKLRKEFSPFGSITSAKVMLE--EGRSKGFGFV 338
Query 61 DFAEPLAAAKCVACLAGRKLHGRSLVVNCNEKVRAKIAKVTEERI--LATMRAYKNKT 116
F+ P A K V + GR + + L V ++ + A +T + + +A MRA T
Sbjct 339 CFSSPEEATKAVTEMNGRIVGSKPLYVALAQRKEERKAHLTNQYMQRIAGMRALPANT 396
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 22/79 (27%), Positives = 40/79 (50%), Gaps = 1/79 (1%)
Query 14 TTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVA 73
T +YI N + D++ ++ ++G+ L +DP SG+ FGF F A K V
Sbjct 191 TNVYIKNFGEDMDDERLKETFSKYGKTLSVKVMTDP-SGKSKGFGFVSFERHEDANKAVD 249
Query 74 CLAGRKLHGRSLVVNCNEK 92
+ G+ ++G+ + V +K
Sbjct 250 DMNGKDVNGKIMFVGRAQK 268
> ath:AT3G52150 RNA recognition motif (RRM)-containing protein
Length=253
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 26/77 (33%), Positives = 35/77 (45%), Gaps = 0/77 (0%)
Query 16 LYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVACL 75
+YIGNI + N+ + L+ E G V + D SG+ FGF A V L
Sbjct 78 VYIGNIPRTVTNEQLTKLVEEHGAVEKVQVMYDKYSGRSRRFGFATMKSVEDANAVVEKL 137
Query 76 AGRKLHGRSLVVNCNEK 92
G + GR + VN EK
Sbjct 138 NGNTVEGREIKVNITEK 154
Score = 39.3 bits (90), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 19/73 (26%), Positives = 32/73 (43%), Gaps = 0/73 (0%)
Query 16 LYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVACL 75
+Y+GN+ K + + NL E G+V+ P + + FGF F+ + L
Sbjct 179 VYVGNLAKTVTKEMLENLFSEKGKVVSAKVSRVPGTSKSTGFGFVTFSSEEDVEAAIVAL 238
Query 76 AGRKLHGRSLVVN 88
L G+ + VN
Sbjct 239 NNSLLEGQKIRVN 251
> mmu:170791 Rbm39, 1500012C14Rik, 2310040E03Rik, B330012G18Rik,
C79248, R75070, Rnpc2, caper; RNA binding motif protein 39;
K13091 RNA-binding protein 39
Length=530
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 21/73 (28%), Positives = 36/73 (49%), Gaps = 0/73 (0%)
Query 16 LYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVACL 75
LY+G++ + D +R + FGR+ D +G+ +GF F++ A K + L
Sbjct 252 LYVGSLHFNITEDMLRGIFEPFGRIESIQLMMDSETGRSKGYGFITFSDSECAKKALEQL 311
Query 76 AGRKLHGRSLVVN 88
G +L GR + V
Sbjct 312 NGFELAGRPMKVG 324
> dre:321035 pabpc4, cb12, sb:cb12; poly(A) binding protein, cytoplasmic
4 (inducible form); K13126 polyadenylate-binding
protein
Length=637
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 26/99 (26%), Positives = 50/99 (50%), Gaps = 8/99 (8%)
Query 14 TTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVA 73
T +YI N D+ ++ L ++G+ L +DP +G+ FGF + + A K V
Sbjct 192 TNVYIKNFGDDMDDQRLKELFDKYGKTLSVKVMTDP-TGKSRGFGFVSYEKHEDANKAVE 250
Query 74 CLAGRKLHGRSLVVNCNEK-------VRAKIAKVTEERI 105
+ G +L+G+++ V +K ++ K ++ +ERI
Sbjct 251 EMNGTELNGKTVFVGRAQKKMERQAELKRKFEQLKQERI 289
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 29/113 (25%), Positives = 53/113 (46%), Gaps = 4/113 (3%)
Query 1 QQQQQQQQQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFC 60
+Q +Q++ + + LYI N+ D++ +R FG + + G+ FGF
Sbjct 282 EQLKQERISRYQGVNLYIKNLDDTIDDEKLRKEFSPFGSITSAKVMLE--EGRSKGFGFV 339
Query 61 DFAEPLAAAKCVACLAGRKLHGRSLVVNCNEKVRAKIAKVTEERI--LATMRA 111
F+ P A K V + GR + + L V ++ + A +T + + +A MRA
Sbjct 340 CFSSPEEATKAVTEMNGRIVGSKPLYVALAQRKEERKAHLTNQYMQRIAGMRA 392
> hsa:9584 RBM39, CAPER, CAPERalpha, CC1.3, DKFZp781C0423, FLJ44170,
HCC1, RNPC2, fSAP59; RNA binding motif protein 39; K13091
RNA-binding protein 39
Length=524
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 21/73 (28%), Positives = 36/73 (49%), Gaps = 0/73 (0%)
Query 16 LYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVACL 75
LY+G++ + D +R + FGR+ D +G+ +GF F++ A K + L
Sbjct 252 LYVGSLHFNITEDMLRGIFEPFGRIESIQLMMDSETGRSKGYGFITFSDSECAKKALEQL 311
Query 76 AGRKLHGRSLVVN 88
G +L GR + V
Sbjct 312 NGFELAGRPMKVG 324
Lambda K H
0.316 0.131 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3321543300
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40