bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2827_orf1
Length=102
Score E
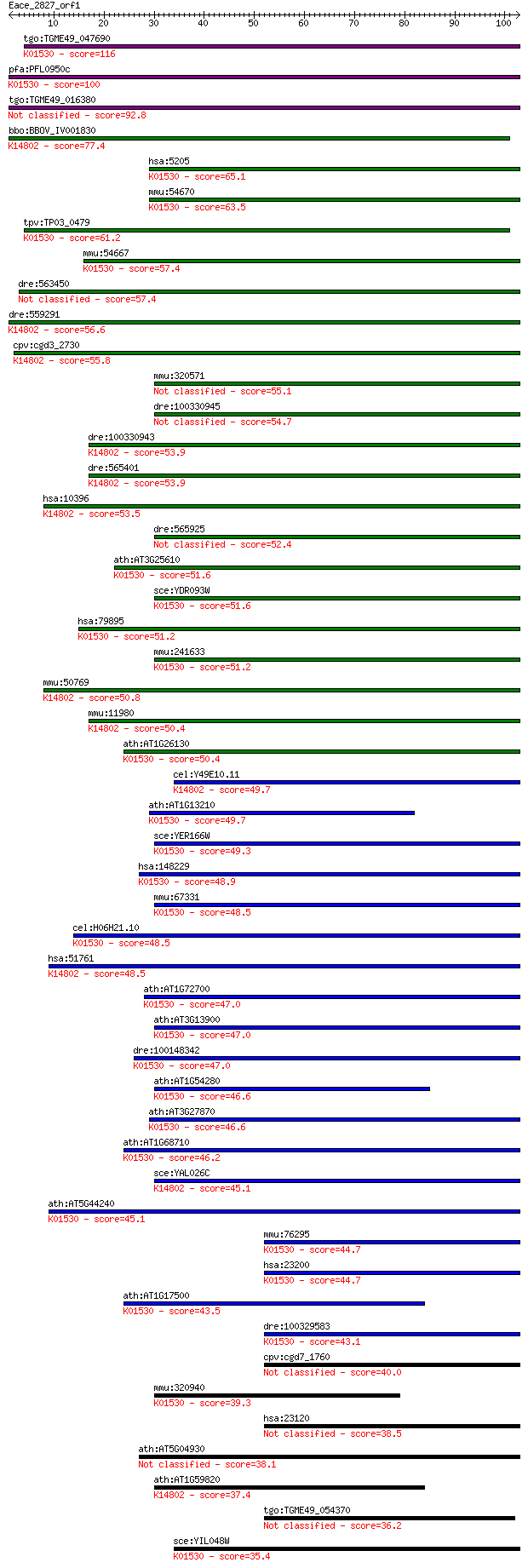
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_047690 p-type ATPase2, putative (EC:3.6.3.1); K0153... 116 1e-26
pfa:PFL0950c ATPase2; aminophospholipid-transporting P-ATPase ... 100 2e-21
tgo:TGME49_016380 ATPase 2, putative (EC:3.6.3.1 3.6.3.9) 92.8 2e-19
bbo:BBOV_IV001830 21.m02975; phospholipid-translocating P-type... 77.4 1e-14
hsa:5205 ATP8B1, ATPIC, BRIC, FIC1, PFIC, PFIC1; ATPase, amino... 65.1 5e-11
mmu:54670 Atp8b1, AI451886, FIC1, Ic; ATPase, class I, type 8B... 63.5 1e-10
tpv:TP03_0479 P-type ATPase (EC:3.6.3.-); K01530 phospholipid-... 61.2 8e-10
mmu:54667 Atp8b2, Id; ATPase, class I, type 8B, member 2 (EC:3... 57.4 1e-08
dre:563450 similar to Potential phospholipid-transporting ATPa... 57.4 1e-08
dre:559291 ATPase, class I, type 8B, member 2-like; K14802 pho... 56.6 2e-08
cpv:cgd3_2730 protein with 10 transmembrane domains, possible ... 55.8 3e-08
mmu:320571 Atp8b5, 4930417M19Rik, Feta; ATPase, class I, type ... 55.1 5e-08
dre:100330945 ATPase, class I, type 8B, member 1-like 54.7 7e-08
dre:100330943 atp8a2; ATPase, aminophospholipid transporter, c... 53.9 1e-07
dre:565401 ATPase, aminophospholipid transporter-like, class I... 53.9 1e-07
hsa:10396 ATP8A1, ATPASEII, ATPIA, ATPP2, MGC130042, MGC130043... 53.5 2e-07
dre:565925 ATPase, aminophospholipid transporter-like, class I... 52.4 3e-07
ath:AT3G25610 haloacid dehalogenase-like hydrolase family prot... 51.6 6e-07
sce:YDR093W DNF2; Aminophospholipid translocase (flippase) tha... 51.6 6e-07
hsa:79895 ATP8B4, ATPIM, FLJ25418; ATPase, class I, type 8B, m... 51.2 7e-07
mmu:241633 Atp8b4, A530043E15, Im, MGC161341; ATPase, class I,... 51.2 9e-07
mmu:50769 Atp8a2, AI415030, Atpc1b, Ib; ATPase, aminophospholi... 50.8 9e-07
mmu:11980 Atp8a1, AI481521, AI853962, APLT, AW743152, AW822227... 50.4 1e-06
ath:AT1G26130 haloacid dehalogenase-like hydrolase family prot... 50.4 1e-06
cel:Y49E10.11 tat-1; Transbilayer Amphipath Transporters (subf... 49.7 2e-06
ath:AT1G13210 ACA.l; ACA.l (autoinhibited Ca2+/ATPase II); ATP... 49.7 2e-06
sce:YER166W DNF1; Aminophospholipid translocase (flippase) tha... 49.3 3e-06
hsa:148229 ATP8B3, ATPIK; ATPase, aminophospholipid transporte... 48.9 4e-06
mmu:67331 Atp8b3, 1700042F02Rik, 1700056N23Rik, ATPIK, SAPLT; ... 48.5 5e-06
cel:H06H21.10 tat-2; Transbilayer Amphipath Transporters (subf... 48.5 5e-06
hsa:51761 ATP8A2, ATP, ATPIB, DKFZp434B1913, IB, ML-1; ATPase,... 48.5 5e-06
ath:AT1G72700 haloacid dehalogenase-like hydrolase family prot... 47.0 1e-05
ath:AT3G13900 ATPase, coupled to transmembrane movement of ion... 47.0 2e-05
dre:100148342 atp8b1; ATPase, aminophospholipid transporter, c... 47.0 2e-05
ath:AT1G54280 haloacid dehalogenase-like hydrolase family prot... 46.6 2e-05
ath:AT3G27870 haloacid dehalogenase-like hydrolase family prot... 46.6 2e-05
ath:AT1G68710 haloacid dehalogenase-like hydrolase family prot... 46.2 2e-05
sce:YAL026C DRS2, FUN38, SWA3; Aminophospholipid translocase (... 45.1 6e-05
ath:AT5G44240 haloacid dehalogenase-like hydrolase family prot... 45.1 6e-05
mmu:76295 Atp11b, 1110019I14Rik, ATPIF, ATPIR, mKIAA0956; ATPa... 44.7 6e-05
hsa:23200 ATP11B, ATPIF, ATPIR, DKFZp434J238, DKFZp434N1615, K... 44.7 8e-05
ath:AT1G17500 ATPase, coupled to transmembrane movement of ion... 43.5 2e-04
dre:100329583 si:dkey-211e20.10; K01530 phospholipid-transloca... 43.1 2e-04
cpv:cgd7_1760 P-type ATpase (calcium/phospholipid-transporter)... 40.0 0.002
mmu:320940 Atp11c, A330005H02Rik, AI315324, Ig, MGC117487; ATP... 39.3 0.003
hsa:23120 ATP10B, ATPVB, FLJ21477, KIAA0715; ATPase, class V, ... 38.5 0.006
ath:AT5G04930 ALA1; ALA1 (aminophospholipid ATPase1); ATPase, ... 38.1 0.006
ath:AT1G59820 ALA3; ALA3 (Aminophospholipid ATPase3); ATPase, ... 37.4 0.010
tgo:TGME49_054370 adenylate and guanylate cyclase catalytic do... 36.2 0.025
sce:YIL048W NEO1; Neo1p (EC:3.6.3.1); K01530 phospholipid-tran... 35.4 0.044
> tgo:TGME49_047690 p-type ATPase2, putative (EC:3.6.3.1); K01530
phospholipid-translocating ATPase [EC:3.6.3.1]
Length=1871
Score = 116 bits (291), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 58/99 (58%), Positives = 69/99 (69%), Gaps = 0/99 (0%)
Query 4 HVQISDARLREHLLDATNPLHAHAVRFFFHLAVNHSVICETAEDGGATYGASSPDEGALV 63
VQ+ D+ LR L D +P+H + V FF HLA+NH+V+ ET G Y ASSPDEGALV
Sbjct 940 QVQMVDSALRHQLNDPNHPMHPYLVDFFLHLAINHAVVLETDPFGMTRYSASSPDEGALV 999
Query 64 YAAHHFGISFLGRHSEGLFVSVLGRKTSVRVLCSIEFTS 102
Y A HFGI FLG+ GL VSVLGRK VRVL S+EF+S
Sbjct 1000 YGARHFGIEFLGQTPSGLEVSVLGRKLHVRVLASVEFSS 1038
> pfa:PFL0950c ATPase2; aminophospholipid-transporting P-ATPase
(EC:3.6.3.1); K01530 phospholipid-translocating ATPase [EC:3.6.3.1]
Length=1555
Score = 100 bits (248), Expect = 2e-21, Method: Composition-based stats.
Identities = 48/102 (47%), Positives = 64/102 (62%), Gaps = 0/102 (0%)
Query 1 QTPHVQISDARLREHLLDATNPLHAHAVRFFFHLAVNHSVICETAEDGGATYGASSPDEG 60
+TPHV I D + HL D + H + + FF HLA+NH+VICE ++G TY +SSPDE
Sbjct 712 KTPHVNIIDNDIINHLKDPNHFNHVNLINFFLHLAINHAVICEKDKEGVTTYSSSSPDEE 771
Query 61 ALVYAAHHFGISFLGRHSEGLFVSVLGRKTSVRVLCSIEFTS 102
ALV AA HF I+FL R +S+ G+ + L +IEFTS
Sbjct 772 ALVNAAKHFDITFLYRREGKYGISIFGKIYEIDTLATIEFTS 813
> tgo:TGME49_016380 ATPase 2, putative (EC:3.6.3.1 3.6.3.9)
Length=1791
Score = 92.8 bits (229), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 48/103 (46%), Positives = 63/103 (61%), Gaps = 1/103 (0%)
Query 1 QTPHVQISDARLREHLLDATNPLHAHAVRFFFHLAVNHSVICETAEDGG-ATYGASSPDE 59
+ PHV++ D LR L D +P+H+H + FF HLAVNHS + E E G Y A+SPDE
Sbjct 827 EAPHVKVGDPTLRAALSDPDDPMHSHIIDFFLHLAVNHSALAEHDEQTGHLCYSANSPDE 886
Query 60 GALVYAAHHFGISFLGRHSEGLFVSVLGRKTSVRVLCSIEFTS 102
A V AA HFG++F R G+ ++VLGR V +L S F S
Sbjct 887 RAFVCAARHFGVTFRARTPFGVELNVLGRTVHVDILASFPFES 929
> bbo:BBOV_IV001830 21.m02975; phospholipid-translocating P-type
ATPase; K14802 phospholipid-transporting ATPase [EC:3.6.3.1]
Length=1098
Score = 77.4 bits (189), Expect = 1e-14, Method: Composition-based stats.
Identities = 37/100 (37%), Positives = 53/100 (53%), Gaps = 0/100 (0%)
Query 1 QTPHVQISDARLREHLLDATNPLHAHAVRFFFHLAVNHSVICETAEDGGATYGASSPDEG 60
+ HV + D L + L ++P H H V FF HL N++V+ + E G Y + SPDE
Sbjct 433 KASHVNLVDDELFKQLKTPSDPRHTHLVEFFMHLVCNNAVLTDIQESGDVQYNSQSPDEL 492
Query 61 ALVYAAHHFGISFLGRHSEGLFVSVLGRKTSVRVLCSIEF 100
V+AA + + S + +SV GRK VR L +IEF
Sbjct 493 CFVHAAKFADFKLMDKTSNSITLSVFGRKMHVRTLANIEF 532
> hsa:5205 ATP8B1, ATPIC, BRIC, FIC1, PFIC, PFIC1; ATPase, aminophospholipid
transporter, class I, type 8B, member 1 (EC:3.6.3.1);
K01530 phospholipid-translocating ATPase [EC:3.6.3.1]
Length=1251
Score = 65.1 bits (157), Expect = 5e-11, Method: Composition-based stats.
Identities = 34/74 (45%), Positives = 47/74 (63%), Gaps = 1/74 (1%)
Query 29 RFFFHLAVNHSVICETAEDGGATYGASSPDEGALVYAAHHFGISFLGRHSEGLFVSVLGR 88
+FFF LAV H+V+ + DG Y A+SPDEGALV AA +FG +FL R + +S LG
Sbjct 526 QFFFLLAVCHTVMVDRT-DGQLNYQAASPDEGALVNAARNFGFAFLARTQNTITISELGT 584
Query 89 KTSVRVLCSIEFTS 102
+ + VL ++F S
Sbjct 585 ERTYNVLAILDFNS 598
> mmu:54670 Atp8b1, AI451886, FIC1, Ic; ATPase, class I, type
8B, member 1 (EC:3.6.3.1); K01530 phospholipid-translocating
ATPase [EC:3.6.3.1]
Length=1251
Score = 63.5 bits (153), Expect = 1e-10, Method: Composition-based stats.
Identities = 33/74 (44%), Positives = 47/74 (63%), Gaps = 1/74 (1%)
Query 29 RFFFHLAVNHSVICETAEDGGATYGASSPDEGALVYAAHHFGISFLGRHSEGLFVSVLGR 88
+FFF L++ H+V+ + DG Y A+SPDEGALV AA +FG +FL R + VS LG
Sbjct 526 QFFFLLSICHTVMVDRI-DGQINYQAASPDEGALVNAARNFGFAFLARTQNTITVSELGS 584
Query 89 KTSVRVLCSIEFTS 102
+ + VL ++F S
Sbjct 585 ERTYNVLAILDFNS 598
> tpv:TP03_0479 P-type ATPase (EC:3.6.3.-); K01530 phospholipid-translocating
ATPase [EC:3.6.3.1]
Length=1405
Score = 61.2 bits (147), Expect = 8e-10, Method: Composition-based stats.
Identities = 35/97 (36%), Positives = 52/97 (53%), Gaps = 2/97 (2%)
Query 4 HVQISDARLREHLLDATNPLHAHAVRFFFHLAVNHSVICETAEDGGATYGASSPDEGALV 63
HV + D L + L D ++P H + V FFFHLA+N+ I ++++D Y SPDE V
Sbjct 690 HVNLVDDTLFQELNDPSHPRHEYLVDFFFHLALNNWAIPDSSDD--RIYMCPSPDELCFV 747
Query 64 YAAHHFGISFLGRHSEGLFVSVLGRKTSVRVLCSIEF 100
AA G L R+S VSV + V+++ +F
Sbjct 748 NAAAFCGFRLLQRNSNFATVSVFNKIYKVKIIAQADF 784
> mmu:54667 Atp8b2, Id; ATPase, class I, type 8B, member 2 (EC:3.6.3.1);
K01530 phospholipid-translocating ATPase [EC:3.6.3.1]
Length=1214
Score = 57.4 bits (137), Expect = 1e-08, Method: Composition-based stats.
Identities = 32/87 (36%), Positives = 47/87 (54%), Gaps = 0/87 (0%)
Query 16 LLDATNPLHAHAVRFFFHLAVNHSVICETAEDGGATYGASSPDEGALVYAAHHFGISFLG 75
LL+A H FF L++ H+V+ E +G Y A SPDEGALV AA +FG F
Sbjct 453 LLEAVKMGDPHTHEFFRLLSLCHTVMSEEKNEGELYYKAQSPDEGALVTAARNFGFVFRS 512
Query 76 RHSEGLFVSVLGRKTSVRVLCSIEFTS 102
R + + V LG + ++L ++F +
Sbjct 513 RTPKTITVHELGTAITYQLLAILDFNN 539
> dre:563450 similar to Potential phospholipid-transporting ATPase
ID (ATPase class I type 8B member 2)
Length=1189
Score = 57.4 bits (137), Expect = 1e-08, Method: Composition-based stats.
Identities = 36/100 (36%), Positives = 52/100 (52%), Gaps = 4/100 (4%)
Query 3 PHVQISDARLREHLLDATNPLHAHAVRFFFHLAVNHSVICETAEDGGATYGASSPDEGAL 62
P D +L E + + +HA FF LA+ H+V+ E G Y A SPDEGAL
Sbjct 451 PKFFFHDHKLVEAVKLGSPEVHA----FFRLLALCHTVMPEEKTQGDLFYQAQSPDEGAL 506
Query 63 VYAAHHFGISFLGRHSEGLFVSVLGRKTSVRVLCSIEFTS 102
V AA +FG F R E + V +G +T+ +L ++F +
Sbjct 507 VTAARNFGFVFRARTPETISVVEMGIETTYELLAVLDFNN 546
> dre:559291 ATPase, class I, type 8B, member 2-like; K14802 phospholipid-transporting
ATPase [EC:3.6.3.1]
Length=1223
Score = 56.6 bits (135), Expect = 2e-08, Method: Composition-based stats.
Identities = 37/110 (33%), Positives = 56/110 (50%), Gaps = 8/110 (7%)
Query 1 QTPHVQIS-----DARLREH---LLDATNPLHAHAVRFFFHLAVNHSVICETAEDGGATY 52
+TP V S D + R H L++A FF LA+ H+V+ E +G Y
Sbjct 459 KTPCVDFSFNPLMDRKFRFHDSSLVEAIKLEEPLVQEFFRLLALCHTVMPEERNEGELVY 518
Query 53 GASSPDEGALVYAAHHFGISFLGRHSEGLFVSVLGRKTSVRVLCSIEFTS 102
A SPDEGALV AA +FG F R E + + +G+ + ++L ++F +
Sbjct 519 QAQSPDEGALVTAARNFGFVFRSRTPETITLYEMGQAVTYQLLAILDFNN 568
> cpv:cgd3_2730 protein with 10 transmembrane domains, possible
calcium transporting ATpase or aminophospholipid transporter
; K14802 phospholipid-transporting ATPase [EC:3.6.3.1]
Length=1178
Score = 55.8 bits (133), Expect = 3e-08, Method: Composition-based stats.
Identities = 35/104 (33%), Positives = 54/104 (51%), Gaps = 10/104 (9%)
Query 2 TPHVQISDARLREHLLDATNPLHAHAVRFFFHLAVNHSVICETAE---DGGATYGASSPD 58
TPHV + D LR++L + + + + A+NH+V+ + E D Y ASS D
Sbjct 454 TPHVDMEDELLRKNLKEG----NKEEIEILLNFALNHTVLIQENEVDLDEPPLYSASSTD 509
Query 59 EGALVYAAHHFGISFLGRHSEGLFVSVLGRKTSVRVLCSIEFTS 102
E ALV A+HHFGIS + ++ + G T+ S+EF +
Sbjct 510 EEALVLASHHFGISLVSQNCSKSTIKFSGIGTN---QTSVEFET 550
> mmu:320571 Atp8b5, 4930417M19Rik, Feta; ATPase, class I, type
8B, member 5 (EC:3.6.3.1)
Length=1183
Score = 55.1 bits (131), Expect = 5e-08, Method: Composition-based stats.
Identities = 28/73 (38%), Positives = 42/73 (57%), Gaps = 0/73 (0%)
Query 30 FFFHLAVNHSVICETAEDGGATYGASSPDEGALVYAAHHFGISFLGRHSEGLFVSVLGRK 89
FF L++ H+V+ E +G Y A SPDEGALV A +FG F R E + V +G+
Sbjct 490 FFLCLSLCHTVMSEEKVEGELVYQAQSPDEGALVTATRNFGFVFCSRTPETITVIEMGKI 549
Query 90 TSVRVLCSIEFTS 102
R+L ++F++
Sbjct 550 RVYRLLAILDFSN 562
> dre:100330945 ATPase, class I, type 8B, member 1-like
Length=1695
Score = 54.7 bits (130), Expect = 7e-08, Method: Composition-based stats.
Identities = 27/73 (36%), Positives = 42/73 (57%), Gaps = 0/73 (0%)
Query 30 FFFHLAVNHSVICETAEDGGATYGASSPDEGALVYAAHHFGISFLGRHSEGLFVSVLGRK 89
FF LA+ H+ + E ++G Y A SPDEGALV AA +FG F R E + + +G +
Sbjct 960 FFRLLALCHTCMAEEKKEGHLVYQAQSPDEGALVTAARNFGFVFRSRSPETITIEEMGIQ 1019
Query 90 TSVRVLCSIEFTS 102
+ +L ++F +
Sbjct 1020 RTYELLAILDFNN 1032
> dre:100330943 atp8a2; ATPase, aminophospholipid transporter,
class I, type 8A, member 2; K14802 phospholipid-transporting
ATPase [EC:3.6.3.1]
Length=889
Score = 53.9 bits (128), Expect = 1e-07, Method: Composition-based stats.
Identities = 29/86 (33%), Positives = 43/86 (50%), Gaps = 1/86 (1%)
Query 17 LDATNPLHAHAVRFFFHLAVNHSVICETAEDGGATYGASSPDEGALVYAAHHFGISFLGR 76
++ +P F +AV H+V+ E ED Y ASSPDEGALV A G F R
Sbjct 174 IEKNHPTSPQICEFLTMMAVCHTVVPER-EDNQIIYQASSPDEGALVKGAKSLGFVFTAR 232
Query 77 HSEGLFVSVLGRKTSVRVLCSIEFTS 102
+ + G++ + +L +EF+S
Sbjct 233 TPHSVIIEARGKEQTYELLNVLEFSS 258
> dre:565401 ATPase, aminophospholipid transporter-like, class
I, type 8A, member 2-like; K14802 phospholipid-transporting
ATPase [EC:3.6.3.1]
Length=916
Score = 53.9 bits (128), Expect = 1e-07, Method: Composition-based stats.
Identities = 29/86 (33%), Positives = 43/86 (50%), Gaps = 1/86 (1%)
Query 17 LDATNPLHAHAVRFFFHLAVNHSVICETAEDGGATYGASSPDEGALVYAAHHFGISFLGR 76
++ +P F +AV H+V+ E ED Y ASSPDEGALV A G F R
Sbjct 507 IEKNHPTSPQICEFLTMMAVCHTVVPER-EDNQIIYQASSPDEGALVKGAKSLGFVFTAR 565
Query 77 HSEGLFVSVLGRKTSVRVLCSIEFTS 102
+ + G++ + +L +EF+S
Sbjct 566 TPHSVIIEARGKEQTYELLNVLEFSS 591
> hsa:10396 ATP8A1, ATPASEII, ATPIA, ATPP2, MGC130042, MGC130043,
MGC26327; ATPase, aminophospholipid transporter (APLT),
class I, type 8A, member 1 (EC:3.6.3.1); K14802 phospholipid-transporting
ATPase [EC:3.6.3.1]
Length=1149
Score = 53.5 bits (127), Expect = 2e-07, Method: Composition-based stats.
Identities = 34/95 (35%), Positives = 49/95 (51%), Gaps = 2/95 (2%)
Query 8 SDARLREHLLDATNPLHAHAVRFFFHLAVNHSVICETAEDGGATYGASSPDEGALVYAAH 67
SD+ L E+L + +P F +AV H+ + E D Y A+SPDEGALV AA
Sbjct 444 SDSSLLENLQN-NHPTAPIICEFLTMMAVCHTAVPEREGDK-IIYQAASPDEGALVRAAK 501
Query 68 HFGISFLGRHSEGLFVSVLGRKTSVRVLCSIEFTS 102
F GR + + + LG++ +L +EFTS
Sbjct 502 QLNFVFTGRTPDSVIIDSLGQEERYELLNVLEFTS 536
> dre:565925 ATPase, aminophospholipid transporter-like, class
I, type 8A, member 2-like
Length=1203
Score = 52.4 bits (124), Expect = 3e-07, Method: Composition-based stats.
Identities = 31/73 (42%), Positives = 44/73 (60%), Gaps = 1/73 (1%)
Query 30 FFFHLAVNHSVICETAEDGGATYGASSPDEGALVYAAHHFGISFLGRHSEGLFVSVLGRK 89
FF LA+ H+V+ E +DG Y A+SPDE ALV AA G FL R + L +S +G
Sbjct 466 FFTALALCHTVMSEW-KDGLPHYQAASPDEEALVCAARELGWVFLSRTRDTLTISEMGLT 524
Query 90 TSVRVLCSIEFTS 102
+ ++L ++FTS
Sbjct 525 RNYQLLALLDFTS 537
> ath:AT3G25610 haloacid dehalogenase-like hydrolase family protein;
K01530 phospholipid-translocating ATPase [EC:3.6.3.1]
Length=1202
Score = 51.6 bits (122), Expect = 6e-07, Method: Composition-based stats.
Identities = 36/88 (40%), Positives = 43/88 (48%), Gaps = 7/88 (7%)
Query 22 PLHAHAVRFFFHLAVNHSVICETAEDGG-ATYGASSPDEGALVYAAHHFGISFLGRHSEG 80
P A +FF LAV H+ I ET E+ G +Y A SPDE A V AA FG F R G
Sbjct 507 PEAAVLQKFFRLLAVCHTAIPETDEESGNVSYEAESPDEAAFVVAAREFGFEFFNRTQNG 566
Query 81 LFVSVL----GRKTS--VRVLCSIEFTS 102
+ L G K R+L +EF S
Sbjct 567 ISFRELDLVSGEKVERVYRLLNVLEFNS 594
> sce:YDR093W DNF2; Aminophospholipid translocase (flippase) that
localizes primarily to the plasma membrane; contributes
to endocytosis, protein transport and cell polarity; type 4
P-type ATPase (EC:3.6.3.1); K01530 phospholipid-translocating
ATPase [EC:3.6.3.1]
Length=1612
Score = 51.6 bits (122), Expect = 6e-07, Method: Composition-based stats.
Identities = 30/75 (40%), Positives = 38/75 (50%), Gaps = 2/75 (2%)
Query 30 FFFHLAVNHSVICETAEDGGATYG--ASSPDEGALVYAAHHFGISFLGRHSEGLFVSVLG 87
F LA+ HSV+ E +D A SPDE ALV A G SF+G GL V + G
Sbjct 815 FLLALALCHSVLVEPNKDDPKKLDIKAQSPDESALVSTARQLGYSFVGSSKSGLIVEIQG 874
Query 88 RKTSVRVLCSIEFTS 102
+ +VL +EF S
Sbjct 875 VQKEFQVLNVLEFNS 889
> hsa:79895 ATP8B4, ATPIM, FLJ25418; ATPase, class I, type 8B,
member 4 (EC:3.6.3.1); K01530 phospholipid-translocating ATPase
[EC:3.6.3.1]
Length=1192
Score = 51.2 bits (121), Expect = 7e-07, Method: Composition-based stats.
Identities = 29/88 (32%), Positives = 44/88 (50%), Gaps = 0/88 (0%)
Query 15 HLLDATNPLHAHAVRFFFHLAVNHSVICETAEDGGATYGASSPDEGALVYAAHHFGISFL 74
HL+++ F LA+ H+V+ E G Y SPDEGALV AA +FG F
Sbjct 452 HLMESIKMGDPKVHEFLRLLALCHTVMSEENSAGELIYQVQSPDEGALVTAARNFGFIFK 511
Query 75 GRHSEGLFVSVLGRKTSVRVLCSIEFTS 102
R E + + LG + ++L ++F +
Sbjct 512 SRTPETITIEELGTLVTYQLLAFLDFNN 539
> mmu:241633 Atp8b4, A530043E15, Im, MGC161341; ATPase, class
I, type 8B, member 4; K01530 phospholipid-translocating ATPase
[EC:3.6.3.1]
Length=1194
Score = 51.2 bits (121), Expect = 9e-07, Method: Composition-based stats.
Identities = 27/73 (36%), Positives = 39/73 (53%), Gaps = 0/73 (0%)
Query 30 FFFHLAVNHSVICETAEDGGATYGASSPDEGALVYAAHHFGISFLGRHSEGLFVSVLGRK 89
F LA+ H+V+ E G Y SPDEGALV AA +FG F R E + + LG
Sbjct 469 FLRLLALCHTVMSEENSAGQLVYQVQSPDEGALVTAARNFGFIFKSRTPETITIEELGTP 528
Query 90 TSVRVLCSIEFTS 102
+ ++L ++F +
Sbjct 529 VTYQLLAFLDFNN 541
> mmu:50769 Atp8a2, AI415030, Atpc1b, Ib; ATPase, aminophospholipid
transporter-like, class I, type 8A, member 2 (EC:3.6.3.1);
K14802 phospholipid-transporting ATPase [EC:3.6.3.1]
Length=1148
Score = 50.8 bits (120), Expect = 9e-07, Method: Composition-based stats.
Identities = 34/96 (35%), Positives = 51/96 (53%), Gaps = 4/96 (4%)
Query 8 SDARLREHLLDATNPLHAHAVRFFFHLAVNHSVICETAEDGGAT-YGASSPDEGALVYAA 66
+D RL +++ D +P F LAV H+V+ E +DG Y ASSPDE ALV A
Sbjct 439 NDPRLLKNIED-QHPTAPCIQEFLTLLAVCHTVVPE--KDGDEIIYQASSPDEAALVKGA 495
Query 67 HHFGISFLGRHSEGLFVSVLGRKTSVRVLCSIEFTS 102
G F GR + + +G++ + +L +EF+S
Sbjct 496 KKLGFVFTGRTPYSVIIEAMGQEQTFGILNVLEFSS 531
> mmu:11980 Atp8a1, AI481521, AI853962, APLT, AW743152, AW822227,
Atp3a2, B230107D19Rik, ClassI, KIAA4233, mKIAA4233; ATPase,
aminophospholipid transporter (APLT), class I, type 8A,
member 1 (EC:3.6.3.1); K14802 phospholipid-transporting ATPase
[EC:3.6.3.1]
Length=1164
Score = 50.4 bits (119), Expect = 1e-06, Method: Composition-based stats.
Identities = 30/86 (34%), Positives = 42/86 (48%), Gaps = 1/86 (1%)
Query 17 LDATNPLHAHAVRFFFHLAVNHSVICETAEDGGATYGASSPDEGALVYAAHHFGISFLGR 76
L +P F +AV H+ + E D Y A+SPDEGALV AA F GR
Sbjct 467 LQNNHPTAPIICEFLTMMAVCHTAVPEREGDK-IIYQAASPDEGALVRAAKQLNFVFTGR 525
Query 77 HSEGLFVSVLGRKTSVRVLCSIEFTS 102
+ + + LG++ +L +EFTS
Sbjct 526 TPDSVIIDSLGQEERYELLNVLEFTS 551
> ath:AT1G26130 haloacid dehalogenase-like hydrolase family protein;
K01530 phospholipid-translocating ATPase [EC:3.6.3.1]
Length=1184
Score = 50.4 bits (119), Expect = 1e-06, Method: Composition-based stats.
Identities = 36/87 (41%), Positives = 43/87 (49%), Gaps = 8/87 (9%)
Query 24 HAHAVRFFFHL-AVNHSVICETAEDGGA-TYGASSPDEGALVYAAHHFGISFLGRHSEGL 81
HA ++ FF L AV H+VI E ED G +Y A SPDE A V AA G F R +
Sbjct 510 HADVIQKFFQLLAVCHTVIPEVDEDTGKISYEAESPDEAAFVIAARELGFEFFTRTQTTI 569
Query 82 FVSVLGRKTSVR------VLCSIEFTS 102
V L T R VL +EF+S
Sbjct 570 SVRELDLVTGERVERLYSVLNVLEFSS 596
> cel:Y49E10.11 tat-1; Transbilayer Amphipath Transporters (subfamily
IV P-type ATPase) family member (tat-1); K14802 phospholipid-transporting
ATPase [EC:3.6.3.1]
Length=1139
Score = 49.7 bits (117), Expect = 2e-06, Method: Composition-based stats.
Identities = 27/69 (39%), Positives = 40/69 (57%), Gaps = 1/69 (1%)
Query 34 LAVNHSVICETAEDGGATYGASSPDEGALVYAAHHFGISFLGRHSEGLFVSVLGRKTSVR 93
+AV H+V+ E +DG Y +SSPDE ALV A +SF R + + +V G ++
Sbjct 444 MAVCHTVVPEN-KDGQLIYQSSSPDEAALVRGAASQSVSFHTRQPQKVICNVFGEDETIE 502
Query 94 VLCSIEFTS 102
+L I+FTS
Sbjct 503 ILDVIDFTS 511
> ath:AT1G13210 ACA.l; ACA.l (autoinhibited Ca2+/ATPase II); ATPase,
coupled to transmembrane movement of ions, phosphorylative
mechanism / calmodulin binding; K01530 phospholipid-translocating
ATPase [EC:3.6.3.1]
Length=1203
Score = 49.7 bits (117), Expect = 2e-06, Method: Composition-based stats.
Identities = 26/54 (48%), Positives = 30/54 (55%), Gaps = 1/54 (1%)
Query 29 RFFFHLAVNHSVICETAE-DGGATYGASSPDEGALVYAAHHFGISFLGRHSEGL 81
+FF LAV H+ I ET E G +Y A SPDE A V AA FG F R G+
Sbjct 513 KFFRLLAVCHTAIPETDEATGSVSYEAESPDEAAFVVAAREFGFEFFSRTQNGI 566
> sce:YER166W DNF1; Aminophospholipid translocase (flippase) that
localizes primarily to the plasma membrane; contributes
to endocytosis, protein transport and cell polarity; type 4
P-type ATPase (EC:3.6.3.1); K01530 phospholipid-translocating
ATPase [EC:3.6.3.1]
Length=1571
Score = 49.3 bits (116), Expect = 3e-06, Method: Composition-based stats.
Identities = 28/75 (37%), Positives = 38/75 (50%), Gaps = 2/75 (2%)
Query 30 FFFHLAVNHSVICETAEDGGATYG--ASSPDEGALVYAAHHFGISFLGRHSEGLFVSVLG 87
F LA+ HSV+ E D A SPDE ALV A G SF+G+ +GL + + G
Sbjct 770 FMLALALCHSVLVEANPDNPKKLDLKAQSPDEAALVATARDVGFSFVGKTKKGLIIEMQG 829
Query 88 RKTSVRVLCSIEFTS 102
+ +L +EF S
Sbjct 830 IQKEFEILNILEFNS 844
> hsa:148229 ATP8B3, ATPIK; ATPase, aminophospholipid transporter,
class I, type 8B, member 3 (EC:3.6.3.1); K01530 phospholipid-translocating
ATPase [EC:3.6.3.1]
Length=1263
Score = 48.9 bits (115), Expect = 4e-06, Method: Composition-based stats.
Identities = 31/80 (38%), Positives = 45/80 (56%), Gaps = 4/80 (5%)
Query 27 AVRFFFHL-AVNHSVICETA---EDGGATYGASSPDEGALVYAAHHFGISFLGRHSEGLF 82
AVR F+ L A+ H+V+ + Y A+SPDEGALV AA +FG FL R + +
Sbjct 513 AVREFWRLLAICHTVMVRESPRERPDQLLYQAASPDEGALVTAARNFGYVFLSRTQDTVT 572
Query 83 VSVLGRKTSVRVLCSIEFTS 102
+ LG + +VL ++F S
Sbjct 573 IMELGEERVYQVLAIMDFNS 592
> mmu:67331 Atp8b3, 1700042F02Rik, 1700056N23Rik, ATPIK, SAPLT;
ATPase, class I, type 8B, member 3 (EC:3.6.3.1); K01530 phospholipid-translocating
ATPase [EC:3.6.3.1]
Length=1335
Score = 48.5 bits (114), Expect = 5e-06, Method: Composition-based stats.
Identities = 28/73 (38%), Positives = 43/73 (58%), Gaps = 1/73 (1%)
Query 30 FFFHLAVNHSVICETAEDGGATYGASSPDEGALVYAAHHFGISFLGRHSEGLFVSVLGRK 89
F+ LA+ H+V+ + +D Y A+SPDE ALV AA +FG FL R + + + LG +
Sbjct 476 FWRLLAICHTVMVQE-KDNQLLYQAASPDEEALVTAARNFGYVFLSRTQDTITLVELGEE 534
Query 90 TSVRVLCSIEFTS 102
+VL ++F S
Sbjct 535 RVYQVLAMMDFNS 547
> cel:H06H21.10 tat-2; Transbilayer Amphipath Transporters (subfamily
IV P-type ATPase) family member (tat-2); K01530 phospholipid-translocating
ATPase [EC:3.6.3.1]
Length=1222
Score = 48.5 bits (114), Expect = 5e-06, Method: Composition-based stats.
Identities = 29/89 (32%), Positives = 49/89 (55%), Gaps = 1/89 (1%)
Query 14 EHLLDATNPLHAHAVRFFFHLAVNHSVICETAEDGGATYGASSPDEGALVYAAHHFGISF 73
++L+DAT +F+ LA+ H+V+ E + G Y A SPDE AL AA +FG F
Sbjct 377 KNLVDATKRQVPEIDQFWRLLALCHTVMPER-DKGQLVYQAQSPDEHALTSAARNFGYVF 435
Query 74 LGRHSEGLFVSVLGRKTSVRVLCSIEFTS 102
R + + + V+G + + +L ++F +
Sbjct 436 RARTPQSITIEVMGNEETHELLAILDFNN 464
> hsa:51761 ATP8A2, ATP, ATPIB, DKFZp434B1913, IB, ML-1; ATPase,
aminophospholipid transporter, class I, type 8A, member 2
(EC:3.6.3.1); K14802 phospholipid-transporting ATPase [EC:3.6.3.1]
Length=1188
Score = 48.5 bits (114), Expect = 5e-06, Method: Composition-based stats.
Identities = 32/94 (34%), Positives = 47/94 (50%), Gaps = 2/94 (2%)
Query 9 DARLREHLLDATNPLHAHAVRFFFHLAVNHSVICETAEDGGATYGASSPDEGALVYAAHH 68
D RL +++ D +P F LAV H+V+ E D Y ASSPDE ALV A
Sbjct 480 DPRLLKNIED-RHPTAPCIQEFLTLLAVCHTVVPEKDGDN-IIYQASSPDEAALVKGAKK 537
Query 69 FGISFLGRHSEGLFVSVLGRKTSVRVLCSIEFTS 102
G F R + + +G++ + +L +EF+S
Sbjct 538 LGFVFTARTPFSVIIEAMGQEQTFGILNVLEFSS 571
> ath:AT1G72700 haloacid dehalogenase-like hydrolase family protein;
K01530 phospholipid-translocating ATPase [EC:3.6.3.1]
Length=1228
Score = 47.0 bits (110), Expect = 1e-05, Method: Composition-based stats.
Identities = 30/84 (35%), Positives = 44/84 (52%), Gaps = 11/84 (13%)
Query 28 VRFFFHLAVNHSVICETAEDGGA-TYGASSPDEGALVYAAHHFGISFLGRHSEGLFV--- 83
++FF LA+ H+ I E E+ G TY A SPDE + + AA FG F R +F+
Sbjct 534 LQFFRILAICHTAIPELNEETGKYTYEAESPDEASFLAAAREFGFEFFKRTQSSVFIRER 593
Query 84 -----SVLGRKTSVRVLCSIEFTS 102
++ R+ +VL +EFTS
Sbjct 594 FSGSGQIIERE--YKVLNLLEFTS 615
> ath:AT3G13900 ATPase, coupled to transmembrane movement of ions,
phosphorylative mechanism; K01530 phospholipid-translocating
ATPase [EC:3.6.3.1]
Length=1243
Score = 47.0 bits (110), Expect = 2e-05, Method: Composition-based stats.
Identities = 32/78 (41%), Positives = 39/78 (50%), Gaps = 5/78 (6%)
Query 30 FFFHLAVNHSVICETAED-GGATYGASSPDEGALVYAAHHFGISFLGRHSEGLFVSVLGR 88
F LAV H+ I E ED G TY A SPDE A + AA FG F R +F+S
Sbjct 545 FLRILAVCHTAIPEVDEDTGKCTYEAESPDEVAFLVAAGEFGFEFTKRTQSSVFISERHS 604
Query 89 KTSV----RVLCSIEFTS 102
V +VL ++FTS
Sbjct 605 GQPVEREYKVLNVLDFTS 622
> dre:100148342 atp8b1; ATPase, aminophospholipid transporter,
class I, type 8B, member 1; K01530 phospholipid-translocating
ATPase [EC:3.6.3.1]
Length=1259
Score = 47.0 bits (110), Expect = 2e-05, Method: Composition-based stats.
Identities = 27/86 (31%), Positives = 43/86 (50%), Gaps = 9/86 (10%)
Query 26 HAVRFFFHLAVNHSVICETAED---------GGATYGASSPDEGALVYAAHHFGISFLGR 76
+ FF L++ H+V+ E E Y A+SPDEGALV AA +FG FL R
Sbjct 517 QVLEFFKLLSLCHTVMVEEKEGEQKKLHTFPSELVYQAASPDEGALVTAARNFGFVFLSR 576
Query 77 HSEGLFVSVLGRKTSVRVLCSIEFTS 102
+ + + + + + +L ++F S
Sbjct 577 TQDTITIQEMDKPQTYTMLALLDFNS 602
> ath:AT1G54280 haloacid dehalogenase-like hydrolase family protein;
K01530 phospholipid-translocating ATPase [EC:3.6.3.1]
Length=1240
Score = 46.6 bits (109), Expect = 2e-05, Method: Composition-based stats.
Identities = 25/56 (44%), Positives = 31/56 (55%), Gaps = 1/56 (1%)
Query 30 FFFHLAVNHSVICETAEDGG-ATYGASSPDEGALVYAAHHFGISFLGRHSEGLFVS 84
FF LAV H+ I E ED G TY A SPDE A + A+ FG F R +F++
Sbjct 545 FFRILAVCHTAIPEVDEDTGMCTYEAESPDEVAFLVASREFGFEFTKRTQSSVFIA 600
> ath:AT3G27870 haloacid dehalogenase-like hydrolase family protein;
K01530 phospholipid-translocating ATPase [EC:3.6.3.1]
Length=1174
Score = 46.6 bits (109), Expect = 2e-05, Method: Composition-based stats.
Identities = 26/75 (34%), Positives = 37/75 (49%), Gaps = 1/75 (1%)
Query 29 RFFFHLAVNHSVICETAEDGGA-TYGASSPDEGALVYAAHHFGISFLGRHSEGLFVSVLG 87
+FF LA+ H+ I + D G TY A SPDE A V A+ G F R + + +
Sbjct 509 KFFRVLAICHTAIPDVNSDTGEITYEAESPDEAAFVIASRELGFEFFSRSQTSISLHEID 568
Query 88 RKTSVRVLCSIEFTS 102
T +L +EF+S
Sbjct 569 HMTVYELLHVLEFSS 583
> ath:AT1G68710 haloacid dehalogenase-like hydrolase family protein;
K01530 phospholipid-translocating ATPase [EC:3.6.3.1]
Length=1200
Score = 46.2 bits (108), Expect = 2e-05, Method: Composition-based stats.
Identities = 34/87 (39%), Positives = 43/87 (49%), Gaps = 8/87 (9%)
Query 24 HAHAVRFFFHL-AVNHSVICETAEDG-GATYGASSPDEGALVYAAHHFGISFLGRHSEGL 81
HA ++ FF L AV H+VI E ED +Y A SPDE A V AA G F R +
Sbjct 515 HADVIQKFFRLLAVCHTVIPEVDEDTEKISYEAESPDEAAFVIAARELGFEFFNRTQTTI 574
Query 82 FVSVL----GRKTS--VRVLCSIEFTS 102
V L G++ +VL +EF S
Sbjct 575 SVRELDLVSGKRVERLYKVLNVLEFNS 601
> sce:YAL026C DRS2, FUN38, SWA3; Aminophospholipid translocase
(flippase) that maintains membrane lipid asymmetry in post-Golgi
secretory vesicles; contributes to clathrin-coated vesicle
formation and endocytosis; mutations in human homolog ATP8B1
result in liver disease (EC:3.6.3.1); K14802 phospholipid-transporting
ATPase [EC:3.6.3.1]
Length=1355
Score = 45.1 bits (105), Expect = 6e-05, Method: Composition-based stats.
Identities = 31/77 (40%), Positives = 39/77 (50%), Gaps = 6/77 (7%)
Query 30 FFFHLAVNHSVICETAEDGGATYGASSPDEGALVYAAHHFGISFLGRHSEGLFVSVL--- 86
F LA H+VI E DG Y A+SPDEGALV G F+ R +G V+VL
Sbjct 626 FLTLLATCHTVIPEFQSDGSIKYQAASPDEGALVQGGADLGYKFIIR--KGNSVTVLLEE 683
Query 87 -GRKTSVRVLCSIEFTS 102
G + ++L EF S
Sbjct 684 TGEEKEYQLLNICEFNS 700
> ath:AT5G44240 haloacid dehalogenase-like hydrolase family protein;
K01530 phospholipid-translocating ATPase [EC:3.6.3.1]
Length=1107
Score = 45.1 bits (105), Expect = 6e-05, Method: Composition-based stats.
Identities = 30/94 (31%), Positives = 46/94 (48%), Gaps = 0/94 (0%)
Query 9 DARLREHLLDATNPLHAHAVRFFFHLAVNHSVICETAEDGGATYGASSPDEGALVYAAHH 68
DA LL+A +RF +A+ ++V+ ++ G Y A S DE ALV AA
Sbjct 409 DALKDAQLLNAITSGSTDVIRFLTVMAICNTVLPVQSKAGDIVYKAQSQDEDALVIAASK 468
Query 69 FGISFLGRHSEGLFVSVLGRKTSVRVLCSIEFTS 102
+ F+G+++ L + G VL +EFTS
Sbjct 469 LHMVFVGKNANLLEIRFNGSVIRYEVLEILEFTS 502
> mmu:76295 Atp11b, 1110019I14Rik, ATPIF, ATPIR, mKIAA0956; ATPase,
class VI, type 11B (EC:3.6.3.1); K01530 phospholipid-translocating
ATPase [EC:3.6.3.1]
Length=1175
Score = 44.7 bits (104), Expect = 6e-05, Method: Composition-based stats.
Identities = 26/51 (50%), Positives = 31/51 (60%), Gaps = 0/51 (0%)
Query 52 YGASSPDEGALVYAAHHFGISFLGRHSEGLFVSVLGRKTSVRVLCSIEFTS 102
Y ASSPDE ALV AA GI F+G E + V VLGR ++L +EF S
Sbjct 523 YYASSPDEKALVEAAARAGIIFVGISEETMEVKVLGRLERYKLLHILEFDS 573
> hsa:23200 ATP11B, ATPIF, ATPIR, DKFZp434J238, DKFZp434N1615,
KIAA0956, MGC46576; ATPase, class VI, type 11B (EC:3.6.3.1);
K01530 phospholipid-translocating ATPase [EC:3.6.3.1]
Length=1177
Score = 44.7 bits (104), Expect = 8e-05, Method: Composition-based stats.
Identities = 24/51 (47%), Positives = 30/51 (58%), Gaps = 0/51 (0%)
Query 52 YGASSPDEGALVYAAHHFGISFLGRHSEGLFVSVLGRKTSVRVLCSIEFTS 102
Y ASSPDE ALV AA GI F+G E + V LG+ ++L +EF S
Sbjct 524 YYASSPDEKALVEAAARIGIVFIGNSEETMEVKTLGKLERYKLLHILEFDS 574
> ath:AT1G17500 ATPase, coupled to transmembrane movement of ions,
phosphorylative mechanism; K01530 phospholipid-translocating
ATPase [EC:3.6.3.1]
Length=1216
Score = 43.5 bits (101), Expect = 2e-04, Method: Composition-based stats.
Identities = 24/62 (38%), Positives = 32/62 (51%), Gaps = 2/62 (3%)
Query 24 HAHAVRFFFH-LAVNHSVICETAEDGGA-TYGASSPDEGALVYAAHHFGISFLGRHSEGL 81
H + FF LA+ H+ I E E+ G TY A SPDE + + AA FG F R +
Sbjct 518 HTDDILLFFRILAICHTAIPELNEETGKYTYEAESPDEASFLTAASEFGFVFFKRTQSSV 577
Query 82 FV 83
+V
Sbjct 578 YV 579
> dre:100329583 si:dkey-211e20.10; K01530 phospholipid-translocating
ATPase [EC:3.6.3.1]
Length=1149
Score = 43.1 bits (100), Expect = 2e-04, Method: Composition-based stats.
Identities = 20/51 (39%), Positives = 30/51 (58%), Gaps = 0/51 (0%)
Query 52 YGASSPDEGALVYAAHHFGISFLGRHSEGLFVSVLGRKTSVRVLCSIEFTS 102
Y ASSPDE ALV A G++F+G H E + + G+ ++L +EF +
Sbjct 496 YYASSPDEKALVEATKRMGVTFMGSHGEIMEIKKFGKAEKYKLLHVLEFDA 546
> cpv:cgd7_1760 P-type ATpase (calcium/phospholipid-transporter),
9 transmembrane domains
Length=1278
Score = 40.0 bits (92), Expect = 0.002, Method: Composition-based stats.
Identities = 21/53 (39%), Positives = 27/53 (50%), Gaps = 2/53 (3%)
Query 52 YGASSPDEGALVYAAHHFGISF--LGRHSEGLFVSVLGRKTSVRVLCSIEFTS 102
Y A PDE AL Y A + G+S L ++ L + V G V +L I FTS
Sbjct 636 YSAQFPDEAALCYGAQYLGVSLICLNPKTKHLILDVFGELLDVEILAKIPFTS 688
> mmu:320940 Atp11c, A330005H02Rik, AI315324, Ig, MGC117487; ATPase,
class VI, type 11C (EC:3.6.3.1); K01530 phospholipid-translocating
ATPase [EC:3.6.3.1]
Length=1116
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 23/56 (41%), Positives = 30/56 (53%), Gaps = 7/56 (12%)
Query 30 FFFHLAVNHSVICETAED-------GGATYGASSPDEGALVYAAHHFGISFLGRHS 78
F L + H+V +T +D G TY +SSPDE ALV A FG +FLG +
Sbjct 462 FLRALCLCHTVEMKTNDDVDGPVEGAGFTYISSSPDEIALVKGAKRFGFTFLGNQN 517
> hsa:23120 ATP10B, ATPVB, FLJ21477, KIAA0715; ATPase, class V,
type 10B (EC:3.6.3.1)
Length=1461
Score = 38.5 bits (88), Expect = 0.006, Method: Composition-based stats.
Identities = 20/52 (38%), Positives = 30/52 (57%), Gaps = 1/52 (1%)
Query 52 YGASSPDEGALVYAAHHFGISFLGRHSEGLFVSV-LGRKTSVRVLCSIEFTS 102
Y A SPDE ALV+AAH + + + R E + V + G + +LC++ F S
Sbjct 717 YEAESPDEAALVHAAHAYSFTLVSRTPEQVTVRLPQGTCLTFSLLCTLGFDS 768
> ath:AT5G04930 ALA1; ALA1 (aminophospholipid ATPase1); ATPase,
coupled to transmembrane movement of ions, phosphorylative
mechanism
Length=1158
Score = 38.1 bits (87), Expect = 0.006, Method: Composition-based stats.
Identities = 28/81 (34%), Positives = 36/81 (44%), Gaps = 5/81 (6%)
Query 27 AVRFFFHLAVNHSV--ICETAEDGGAT---YGASSPDEGALVYAAHHFGISFLGRHSEGL 81
A FF LA +++ I D Y SPDE ALVYAA +G + R S +
Sbjct 529 ANEFFLSLAACNTIVPIVSNTSDPNVKLVDYQGESPDEQALVYAAAAYGFLLIERTSGHI 588
Query 82 FVSVLGRKTSVRVLCSIEFTS 102
++V G VL EF S
Sbjct 589 VINVRGETQRFNVLGLHEFDS 609
> ath:AT1G59820 ALA3; ALA3 (Aminophospholipid ATPase3); ATPase,
coupled to transmembrane movement of ions, phosphorylative
mechanism / phospholipid transporter; K14802 phospholipid-transporting
ATPase [EC:3.6.3.1]
Length=1213
Score = 37.4 bits (85), Expect = 0.010, Method: Composition-based stats.
Identities = 23/55 (41%), Positives = 30/55 (54%), Gaps = 1/55 (1%)
Query 30 FFFHLAVNHSVICETAED-GGATYGASSPDEGALVYAAHHFGISFLGRHSEGLFV 83
F LA+ H+V+ E E Y A+SPDE ALV AA +FG F R ++V
Sbjct 493 LFRCLAICHTVLPEGDESPEKIVYQAASPDEAALVTAAKNFGFFFYRRTPTMVYV 547
> tgo:TGME49_054370 adenylate and guanylate cyclase catalytic
domain-containing protein (EC:4.6.1.1 3.6.3.1 1.15.1.1 4.6.1.2
3.1.3.48 2.8.3.8)
Length=4368
Score = 36.2 bits (82), Expect = 0.025, Method: Compositional matrix adjust.
Identities = 18/50 (36%), Positives = 29/50 (58%), Gaps = 0/50 (0%)
Query 52 YGASSPDEGALVYAAHHFGISFLGRHSEGLFVSVLGRKTSVRVLCSIEFT 101
Y ASSPDE LV AA H G + + R + +++ G++ +++ EFT
Sbjct 1666 YQASSPDEECLVSAASHMGYTLVSRTNNYAILNINGQERRWQIIGVNEFT 1715
> sce:YIL048W NEO1; Neo1p (EC:3.6.3.1); K01530 phospholipid-translocating
ATPase [EC:3.6.3.1]
Length=1151
Score = 35.4 bits (80), Expect = 0.044, Method: Composition-based stats.
Identities = 26/71 (36%), Positives = 34/71 (47%), Gaps = 3/71 (4%)
Query 34 LAVNHSVICETAEDGGATYGASSPDEGALVYAAHHFGISFL--GRHSEGLFVSVLGRKTS 91
LA+ H+V T ED TY A+SPDE A+V G+S RHS L G+ +
Sbjct 573 LAICHNVT-PTFEDDELTYQAASPDEIAIVKFTESVGLSLFKRDRHSISLLHEHSGKTLN 631
Query 92 VRVLCSIEFTS 102
+L F S
Sbjct 632 YEILQVFPFNS 642
Lambda K H
0.323 0.135 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2031832220
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40