bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2779_orf1
Length=70
Score E
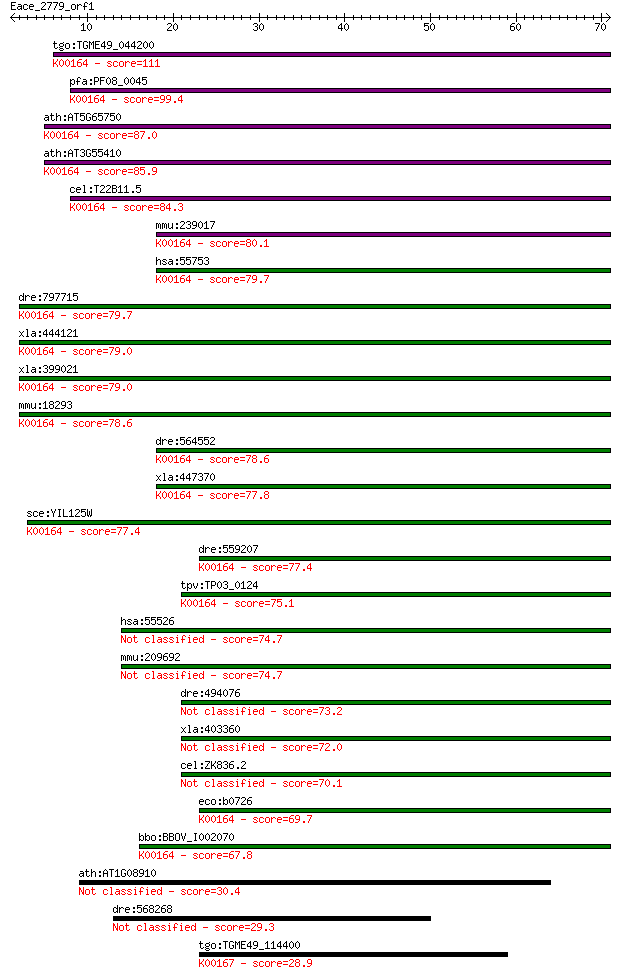
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_044200 2-oxoglutarate dehydrogenase, putative (EC:1... 111 4e-25
pfa:PF08_0045 2-oxoglutarate dehydrogenase E1 component (EC:1.... 99.4 3e-21
ath:AT5G65750 2-oxoglutarate dehydrogenase E1 component, putat... 87.0 1e-17
ath:AT3G55410 2-oxoglutarate dehydrogenase E1 component, putat... 85.9 3e-17
cel:T22B11.5 hypothetical protein; K00164 2-oxoglutarate dehyd... 84.3 8e-17
mmu:239017 Ogdhl; oxoglutarate dehydrogenase-like (EC:1.2.4.-)... 80.1 2e-15
hsa:55753 OGDHL; oxoglutarate dehydrogenase-like (EC:1.2.4.-);... 79.7 2e-15
dre:797715 si:ch211-229p19.3; K00164 2-oxoglutarate dehydrogen... 79.7 2e-15
xla:444121 MGC80496 protein; K00164 2-oxoglutarate dehydrogena... 79.0 3e-15
xla:399021 ogdh, MGC68800, akgdh, e1k, ogdc; oxoglutarate (alp... 79.0 4e-15
mmu:18293 Ogdh, 2210403E04Rik, 2210412K19Rik, AA409584, KIAA41... 78.6 5e-15
dre:564552 ogdh, MGC73296, im:7045267, wu:fa06d01, wu:fb98a04,... 78.6 5e-15
xla:447370 ogdhl, MGC84242; oxoglutarate dehydrogenase-like; K... 77.8 8e-15
sce:YIL125W KGD1, OGD1; Component of the mitochondrial alpha-k... 77.4 1e-14
dre:559207 hypothetical LOC559207; K00164 2-oxoglutarate dehyd... 77.4 1e-14
tpv:TP03_0124 2-oxoglutarate dehydrogenase e1 component (EC:1.... 75.1 5e-14
hsa:55526 DHTKD1, DKFZp762M115, KIAA1630, MGC3090; dehydrogena... 74.7 7e-14
mmu:209692 Dhtkd1, C330018I04Rik; dehydrogenase E1 and transke... 74.7 7e-14
dre:494076 dhtkd1, zgc:101818; dehydrogenase E1 and transketol... 73.2 2e-13
xla:403360 dhtkd1, MGC68840; dehydrogenase E1 and transketolas... 72.0 4e-13
cel:ZK836.2 hypothetical protein 70.1 1e-12
eco:b0726 sucA, ECK0714, JW0715, lys; 2-oxoglutarate decarboxy... 69.7 2e-12
bbo:BBOV_I002070 19.m02351; 2-oxoglutarate dehydrogenase E1 co... 67.8 9e-12
ath:AT1G08910 EMB3001 (embryo defective 3001); zinc ion binding 30.4 1.4
dre:568268 si:ch211-218g4.2 29.3 2.8
tgo:TGME49_114400 branched-chain alpha-keto acid dehydrogenase... 28.9 3.8
> tgo:TGME49_044200 2-oxoglutarate dehydrogenase, putative (EC:1.2.4.2);
K00164 2-oxoglutarate dehydrogenase E1 component
[EC:1.2.4.2]
Length=1116
Score = 111 bits (278), Expect = 4e-25, Method: Composition-based stats.
Identities = 48/65 (73%), Positives = 54/65 (83%), Gaps = 0/65 (0%)
Query 6 HCIFDSLKDLNLPHTINVCNSPLSEYAALGYEFGYSMEHPDALAIWEAQFGDFSNGAQII 65
+ IFDSLK PH I NSPLSEYAA+GYE GYS+EHPD+L IWEAQFGDF+NGAQII
Sbjct 770 YSIFDSLKCYGFPHKIQTVNSPLSEYAAMGYELGYSLEHPDSLCIWEAQFGDFANGAQII 829
Query 66 IDQFV 70
IDQF+
Sbjct 830 IDQFI 834
> pfa:PF08_0045 2-oxoglutarate dehydrogenase E1 component (EC:1.2.4.2);
K00164 2-oxoglutarate dehydrogenase E1 component [EC:1.2.4.2]
Length=1038
Score = 99.4 bits (246), Expect = 3e-21, Method: Composition-based stats.
Identities = 46/63 (73%), Positives = 51/63 (80%), Gaps = 3/63 (4%)
Query 8 IFDSLKDLNLPHTINVCNSPLSEYAALGYEFGYSMEHPDALAIWEAQFGDFSNGAQIIID 67
IFDSLK PHTI V NS LSEYA LGYE GYS EHPDAL IWEAQFGDF+NGAQ++ID
Sbjct 700 IFDSLKT---PHTIEVNNSLLSEYACLGYEIGYSYEHPDALVIWEAQFGDFANGAQVMID 756
Query 68 QFV 70
++
Sbjct 757 NYI 759
> ath:AT5G65750 2-oxoglutarate dehydrogenase E1 component, putative
/ oxoglutarate decarboxylase, putative / alpha-ketoglutaric
dehydrogenase, putative; K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=1025
Score = 87.0 bits (214), Expect = 1e-17, Method: Composition-based stats.
Identities = 36/66 (54%), Positives = 47/66 (71%), Gaps = 0/66 (0%)
Query 5 QHCIFDSLKDLNLPHTINVCNSPLSEYAALGYEFGYSMEHPDALAIWEAQFGDFSNGAQI 64
++C D L P V NS LSE+ LG+E GYSME+P++L IWEAQFGDF+NGAQ+
Sbjct 686 EYCPLDHLIKNQDPEMFTVSNSSLSEFGVLGFELGYSMENPNSLVIWEAQFGDFANGAQV 745
Query 65 IIDQFV 70
+ DQF+
Sbjct 746 MFDQFI 751
> ath:AT3G55410 2-oxoglutarate dehydrogenase E1 component, putative
/ oxoglutarate decarboxylase, putative / alpha-ketoglutaric
dehydrogenase, putative; K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=1017
Score = 85.9 bits (211), Expect = 3e-17, Method: Composition-based stats.
Identities = 36/66 (54%), Positives = 46/66 (69%), Gaps = 0/66 (0%)
Query 5 QHCIFDSLKDLNLPHTINVCNSPLSEYAALGYEFGYSMEHPDALAIWEAQFGDFSNGAQI 64
++C D L P V NS LSE+ LG+E GYSME P++L +WEAQFGDF+NGAQ+
Sbjct 682 EYCPLDHLIMNQDPEMFTVSNSSLSEFGVLGFELGYSMESPNSLVLWEAQFGDFANGAQV 741
Query 65 IIDQFV 70
I DQF+
Sbjct 742 IFDQFI 747
> cel:T22B11.5 hypothetical protein; K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=1029
Score = 84.3 bits (207), Expect = 8e-17, Method: Composition-based stats.
Identities = 40/64 (62%), Positives = 47/64 (73%), Gaps = 1/64 (1%)
Query 8 IFDSLKDLNLPH-TINVCNSPLSEYAALGYEFGYSMEHPDALAIWEAQFGDFSNGAQIII 66
I++ L DL+ VCNS LSEYA LG+E GYSM P++L IWEAQFGDFSN AQ II
Sbjct 699 IYNPLNDLSEGQGEYTVCNSSLSEYAVLGFELGYSMVDPNSLVIWEAQFGDFSNTAQCII 758
Query 67 DQFV 70
DQF+
Sbjct 759 DQFI 762
> mmu:239017 Ogdhl; oxoglutarate dehydrogenase-like (EC:1.2.4.-);
K00164 2-oxoglutarate dehydrogenase E1 component [EC:1.2.4.2]
Length=1029
Score = 80.1 bits (196), Expect = 2e-15, Method: Composition-based stats.
Identities = 35/53 (66%), Positives = 41/53 (77%), Gaps = 2/53 (3%)
Query 18 PHTINVCNSPLSEYAALGYEFGYSMEHPDALAIWEAQFGDFSNGAQIIIDQFV 70
P+T VCNS LSEY LG+E GY+M P+AL +WEAQFGDF N AQ IIDQF+
Sbjct 718 PYT--VCNSSLSEYGVLGFELGYAMASPNALVLWEAQFGDFHNTAQCIIDQFI 768
> hsa:55753 OGDHL; oxoglutarate dehydrogenase-like (EC:1.2.4.-);
K00164 2-oxoglutarate dehydrogenase E1 component [EC:1.2.4.2]
Length=953
Score = 79.7 bits (195), Expect = 2e-15, Method: Composition-based stats.
Identities = 35/53 (66%), Positives = 41/53 (77%), Gaps = 2/53 (3%)
Query 18 PHTINVCNSPLSEYAALGYEFGYSMEHPDALAIWEAQFGDFSNGAQIIIDQFV 70
P+T VCNS LSEY LG+E GY+M P+AL +WEAQFGDF N AQ IIDQF+
Sbjct 642 PYT--VCNSSLSEYGVLGFELGYAMASPNALVLWEAQFGDFHNTAQCIIDQFI 692
> dre:797715 si:ch211-229p19.3; K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=1023
Score = 79.7 bits (195), Expect = 2e-15, Method: Composition-based stats.
Identities = 39/70 (55%), Positives = 49/70 (70%), Gaps = 3/70 (4%)
Query 2 VEAQHCIFDSLKDLN-LPHTINVCNSPLSEYAALGYEFGYSMEHPDALAIWEAQFGDFSN 60
V+ + CI + D N P+T VCNS LSEY LG+E G++M P+AL +WEAQFGDF N
Sbjct 696 VDKRTCIPMNYMDPNQAPYT--VCNSSLSEYGVLGFELGFAMASPNALVLWEAQFGDFHN 753
Query 61 GAQIIIDQFV 70
AQ IIDQF+
Sbjct 754 TAQCIIDQFI 763
> xla:444121 MGC80496 protein; K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=1018
Score = 79.0 bits (193), Expect = 3e-15, Method: Composition-based stats.
Identities = 39/70 (55%), Positives = 48/70 (68%), Gaps = 3/70 (4%)
Query 2 VEAQHCI-FDSLKDLNLPHTINVCNSPLSEYAALGYEFGYSMEHPDALAIWEAQFGDFSN 60
V+ + CI + L P+T VCNS LSEY LG+E G++M P+AL +WEAQFGDF N
Sbjct 691 VDKRTCIPMNHLWPNQAPYT--VCNSSLSEYGVLGFELGFAMASPNALVLWEAQFGDFHN 748
Query 61 GAQIIIDQFV 70
AQ IIDQFV
Sbjct 749 TAQCIIDQFV 758
> xla:399021 ogdh, MGC68800, akgdh, e1k, ogdc; oxoglutarate (alpha-ketoglutarate)
dehydrogenase (lipoamide) (EC:1.2.4.2);
K00164 2-oxoglutarate dehydrogenase E1 component [EC:1.2.4.2]
Length=1021
Score = 79.0 bits (193), Expect = 4e-15, Method: Composition-based stats.
Identities = 39/70 (55%), Positives = 48/70 (68%), Gaps = 3/70 (4%)
Query 2 VEAQHCI-FDSLKDLNLPHTINVCNSPLSEYAALGYEFGYSMEHPDALAIWEAQFGDFSN 60
V+ + CI + L P+T VCNS LSEY LG+E G++M P+AL +WEAQFGDF N
Sbjct 694 VDKRTCIPMNHLWPNQAPYT--VCNSSLSEYGVLGFELGFAMASPNALVLWEAQFGDFHN 751
Query 61 GAQIIIDQFV 70
AQ IIDQFV
Sbjct 752 TAQCIIDQFV 761
> mmu:18293 Ogdh, 2210403E04Rik, 2210412K19Rik, AA409584, KIAA4192,
d1401, mKIAA4192; oxoglutarate dehydrogenase (lipoamide)
(EC:1.2.4.2); K00164 2-oxoglutarate dehydrogenase E1 component
[EC:1.2.4.2]
Length=1023
Score = 78.6 bits (192), Expect = 5e-15, Method: Composition-based stats.
Identities = 38/70 (54%), Positives = 49/70 (70%), Gaps = 3/70 (4%)
Query 2 VEAQHCI-FDSLKDLNLPHTINVCNSPLSEYAALGYEFGYSMEHPDALAIWEAQFGDFSN 60
V+ + CI + L P+T VCNS LSEY LG+E G++M P+AL +WEAQFGDF+N
Sbjct 695 VDKRTCIPMNHLWPNQAPYT--VCNSSLSEYGVLGFELGFAMASPNALVLWEAQFGDFNN 752
Query 61 GAQIIIDQFV 70
AQ IIDQF+
Sbjct 753 MAQCIIDQFI 762
> dre:564552 ogdh, MGC73296, im:7045267, wu:fa06d01, wu:fb98a04,
zgc:73296; oxoglutarate (alpha-ketoglutarate) dehydrogenase
(lipoamide) (EC:1.2.4.2); K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=1022
Score = 78.6 bits (192), Expect = 5e-15, Method: Composition-based stats.
Identities = 34/53 (64%), Positives = 41/53 (77%), Gaps = 2/53 (3%)
Query 18 PHTINVCNSPLSEYAALGYEFGYSMEHPDALAIWEAQFGDFSNGAQIIIDQFV 70
P+T VCNS LSEY LG+E G++M P+AL +WEAQFGDF N AQ IIDQF+
Sbjct 712 PYT--VCNSSLSEYGVLGFELGFAMASPNALVLWEAQFGDFHNTAQCIIDQFI 762
> xla:447370 ogdhl, MGC84242; oxoglutarate dehydrogenase-like;
K00164 2-oxoglutarate dehydrogenase E1 component [EC:1.2.4.2]
Length=1018
Score = 77.8 bits (190), Expect = 8e-15, Method: Composition-based stats.
Identities = 34/53 (64%), Positives = 41/53 (77%), Gaps = 2/53 (3%)
Query 18 PHTINVCNSPLSEYAALGYEFGYSMEHPDALAIWEAQFGDFSNGAQIIIDQFV 70
P+T VCNS LSEY LG+E G++M P+AL +WEAQFGDF N AQ IIDQF+
Sbjct 707 PYT--VCNSSLSEYGVLGFELGFAMASPNALVLWEAQFGDFYNTAQCIIDQFI 757
> sce:YIL125W KGD1, OGD1; Component of the mitochondrial alpha-ketoglutarate
dehydrogenase complex, which catalyzes a key
step in the tricarboxylic acid (TCA) cycle, the oxidative decarboxylation
of alpha-ketoglutarate to form succinyl-CoA (EC:1.2.4.2);
K00164 2-oxoglutarate dehydrogenase E1 component
[EC:1.2.4.2]
Length=1014
Score = 77.4 bits (189), Expect = 1e-14, Method: Composition-based stats.
Identities = 34/69 (49%), Positives = 45/69 (65%), Gaps = 1/69 (1%)
Query 3 EAQHCIFDSLKDLNLPHT-INVCNSPLSEYAALGYEFGYSMEHPDALAIWEAQFGDFSNG 61
+ I+ L LN + NS LSEY +G+E+GYS+ PD L +WEAQFGDF+N
Sbjct 688 QQSEAIYTPLSTLNNEKADFTIANSSLSEYGVMGFEYGYSLTSPDYLVMWEAQFGDFANT 747
Query 62 AQIIIDQFV 70
AQ+IIDQF+
Sbjct 748 AQVIIDQFI 756
> dre:559207 hypothetical LOC559207; K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=1008
Score = 77.4 bits (189), Expect = 1e-14, Method: Composition-based stats.
Identities = 32/48 (66%), Positives = 38/48 (79%), Gaps = 0/48 (0%)
Query 23 VCNSPLSEYAALGYEFGYSMEHPDALAIWEAQFGDFSNGAQIIIDQFV 70
VCNS LSEY LG+E G++M +P+AL WEAQFGDF N AQ IIDQF+
Sbjct 701 VCNSSLSEYGVLGFELGFAMANPNALVCWEAQFGDFHNTAQCIIDQFI 748
> tpv:TP03_0124 2-oxoglutarate dehydrogenase e1 component (EC:1.2.4.2);
K00164 2-oxoglutarate dehydrogenase E1 component [EC:1.2.4.2]
Length=1030
Score = 75.1 bits (183), Expect = 5e-14, Method: Composition-based stats.
Identities = 33/50 (66%), Positives = 40/50 (80%), Gaps = 0/50 (0%)
Query 21 INVCNSPLSEYAALGYEFGYSMEHPDALAIWEAQFGDFSNGAQIIIDQFV 70
+++ NS LSE AALG+E+GYS+ P L IWEAQFGDF NGAQ+IID FV
Sbjct 709 VSIYNSYLSELAALGFEYGYSLYSPKTLNIWEAQFGDFMNGAQVIIDAFV 758
> hsa:55526 DHTKD1, DKFZp762M115, KIAA1630, MGC3090; dehydrogenase
E1 and transketolase domain containing 1 (EC:1.2.4.2)
Length=919
Score = 74.7 bits (182), Expect = 7e-14, Method: Composition-based stats.
Identities = 34/57 (59%), Positives = 40/57 (70%), Gaps = 0/57 (0%)
Query 14 DLNLPHTINVCNSPLSEYAALGYEFGYSMEHPDALAIWEAQFGDFSNGAQIIIDQFV 70
D N + V NSPLSE A LG+E+G S+E P L +WEAQFGDF NGAQII D F+
Sbjct 624 DPNQKGFLEVSNSPLSEEAVLGFEYGMSIESPKLLPLWEAQFGDFFNGAQIIFDTFI 680
> mmu:209692 Dhtkd1, C330018I04Rik; dehydrogenase E1 and transketolase
domain containing 1 (EC:1.2.4.2)
Length=921
Score = 74.7 bits (182), Expect = 7e-14, Method: Composition-based stats.
Identities = 34/57 (59%), Positives = 40/57 (70%), Gaps = 0/57 (0%)
Query 14 DLNLPHTINVCNSPLSEYAALGYEFGYSMEHPDALAIWEAQFGDFSNGAQIIIDQFV 70
D N + V NSPLSE A LG+E+G S+E P L +WEAQFGDF NGAQII D F+
Sbjct 625 DPNQKGFLEVSNSPLSEEAVLGFEYGMSIESPTLLPLWEAQFGDFFNGAQIIFDTFI 681
> dre:494076 dhtkd1, zgc:101818; dehydrogenase E1 and transketolase
domain containing 1 (EC:1.2.4.2)
Length=925
Score = 73.2 bits (178), Expect = 2e-13, Method: Composition-based stats.
Identities = 32/50 (64%), Positives = 37/50 (74%), Gaps = 0/50 (0%)
Query 21 INVCNSPLSEYAALGYEFGYSMEHPDALAIWEAQFGDFSNGAQIIIDQFV 70
+ VCNS LSE A LG+E+G S+ P L IWEAQFGDF NGAQII D F+
Sbjct 637 LEVCNSALSEEAVLGFEYGMSIAQPRLLPIWEAQFGDFFNGAQIIFDTFL 686
> xla:403360 dhtkd1, MGC68840; dehydrogenase E1 and transketolase
domain containing 1 (EC:1.2.4.2)
Length=927
Score = 72.0 bits (175), Expect = 4e-13, Method: Composition-based stats.
Identities = 32/50 (64%), Positives = 37/50 (74%), Gaps = 0/50 (0%)
Query 21 INVCNSPLSEYAALGYEFGYSMEHPDALAIWEAQFGDFSNGAQIIIDQFV 70
+ V NS LSE A LG+E+G S+E P L IWEAQFGDF NGAQII D F+
Sbjct 639 LEVSNSALSEEAVLGFEYGMSIESPKLLPIWEAQFGDFFNGAQIIFDTFI 688
> cel:ZK836.2 hypothetical protein
Length=911
Score = 70.1 bits (170), Expect = 1e-12, Method: Composition-based stats.
Identities = 32/50 (64%), Positives = 39/50 (78%), Gaps = 0/50 (0%)
Query 21 INVCNSPLSEYAALGYEFGYSMEHPDALAIWEAQFGDFSNGAQIIIDQFV 70
+ V N+ LSE A LG+E+G+S E+P L IWEAQFGDF NGAQIIID F+
Sbjct 621 LEVANNLLSEEAILGFEWGFSSENPRRLCIWEAQFGDFFNGAQIIIDTFL 670
> eco:b0726 sucA, ECK0714, JW0715, lys; 2-oxoglutarate decarboxylase,
thiamin-requiring (EC:1.2.4.2); K00164 2-oxoglutarate
dehydrogenase E1 component [EC:1.2.4.2]
Length=933
Score = 69.7 bits (169), Expect = 2e-12, Method: Composition-based stats.
Identities = 29/48 (60%), Positives = 37/48 (77%), Gaps = 0/48 (0%)
Query 23 VCNSPLSEYAALGYEFGYSMEHPDALAIWEAQFGDFSNGAQIIIDQFV 70
V +S LSE A L +E+GY+ P L IWEAQFGDF+NGAQ++IDQF+
Sbjct 654 VWDSVLSEEAVLAFEYGYATAEPRTLTIWEAQFGDFANGAQVVIDQFI 701
> bbo:BBOV_I002070 19.m02351; 2-oxoglutarate dehydrogenase E1
component (EC:1.2.4.2); K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=891
Score = 67.8 bits (164), Expect = 9e-12, Method: Composition-based stats.
Identities = 33/57 (57%), Positives = 41/57 (71%), Gaps = 2/57 (3%)
Query 16 NLPH--TINVCNSPLSEYAALGYEFGYSMEHPDALAIWEAQFGDFSNGAQIIIDQFV 70
N+P+ I V NS LSE AA+ +E+GY +E L IWEAQFGDF+N AQ IID+FV
Sbjct 582 NVPNGSNIEVYNSLLSETAAMAFEYGYGLEDSRVLNIWEAQFGDFANVAQPIIDEFV 638
> ath:AT1G08910 EMB3001 (embryo defective 3001); zinc ion binding
Length=842
Score = 30.4 bits (67), Expect = 1.4, Method: Composition-based stats.
Identities = 21/55 (38%), Positives = 26/55 (47%), Gaps = 2/55 (3%)
Query 9 FDSLKDLNLPHTINVCNSPLSEYAALGYEFGYSMEHPDALAIWEAQFGDFSNGAQ 63
FD + +NLP IN +SP S+ AL F + D LA A FG AQ
Sbjct 453 FDGQQFVNLPQVINTRDSPASQ--ALPMTFSPTPSPQDILATNAANFGTSMPAAQ 505
> dre:568268 si:ch211-218g4.2
Length=964
Score = 29.3 bits (64), Expect = 2.8, Method: Composition-based stats.
Identities = 15/37 (40%), Positives = 20/37 (54%), Gaps = 0/37 (0%)
Query 13 KDLNLPHTINVCNSPLSEYAALGYEFGYSMEHPDALA 49
K L PH ++C+S SEY+ G E M + DA A
Sbjct 144 KLLQFPHNKHICSSSASEYSPAGRECIKHMVNVDATA 180
> tgo:TGME49_114400 branched-chain alpha-keto acid dehydrogenase
E1 component beta chain, putative (EC:1.2.4.1); K00167 2-oxoisovalerate
dehydrogenase E1 component, beta subunit [EC:1.2.4.4]
Length=423
Score = 28.9 bits (63), Expect = 3.8, Method: Composition-based stats.
Identities = 17/36 (47%), Positives = 19/36 (52%), Gaps = 2/36 (5%)
Query 23 VCNSPLSEYAALGYEFGYSMEHPDALAIWEAQFGDF 58
V N+PLSE G FG M AI E QFGD+
Sbjct 151 VFNTPLSEQGIAG--FGIGMAAVGYTAIGEIQFGDY 184
Lambda K H
0.322 0.137 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2024947620
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40