bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2701_orf1
Length=141
Score E
Sequences producing significant alignments: (Bits) Value
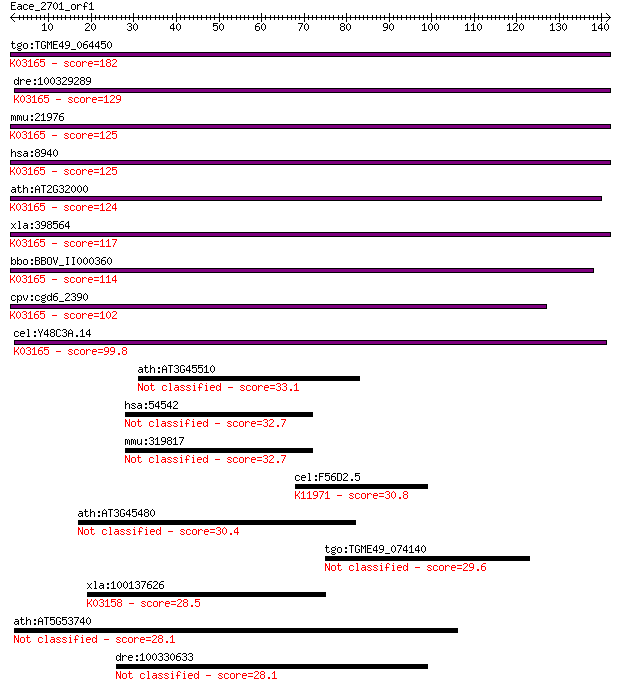
tgo:TGME49_064450 DNA topoisomerase III beta-1, putative (EC:5... 182 2e-46
dre:100329289 topoisomerase (DNA) III beta-like; K03165 DNA to... 129 3e-30
mmu:21976 Top3b; topoisomerase (DNA) III beta (EC:5.99.1.2); K... 125 3e-29
hsa:8940 TOP3B, FLJ39376; topoisomerase (DNA) III beta (EC:5.9... 125 6e-29
ath:AT2G32000 DNA topoisomerase family protein; K03165 DNA top... 124 9e-29
xla:398564 top3b, MGC53016; topoisomerase (DNA) III beta (EC:5... 117 1e-26
bbo:BBOV_II000360 18.m06014; DNA topoisomerase; K03165 DNA top... 114 1e-25
cpv:cgd6_2390 DNA topoisomerase III beta-1 ; K03165 DNA topois... 102 4e-22
cel:Y48C3A.14 hypothetical protein; K03165 DNA topoisomerase I... 99.8 3e-21
ath:AT3G45510 zinc finger (C3HC4-type RING finger) family protein 33.1 0.26
hsa:54542 RC3H2, FLJ20301, FLJ20713, MGC52176, MNAB, RNF164; r... 32.7 0.39
mmu:319817 Rc3h2, 2900024N03Rik, 9430019J22Rik, D930043C02Rik,... 32.7 0.40
cel:F56D2.5 hypothetical protein; K11971 E3 ubiquitin-protein ... 30.8 1.5
ath:AT3G45480 zinc ion binding 30.4 1.7
tgo:TGME49_074140 RNA recognition motif-containing protein 29.6 3.3
xla:100137626 tnfrsf1a, cd120a, fpf, p55, p55-r, tbp1, tnf-r, ... 28.5 7.1
ath:AT5G53740 hypothetical protein 28.1 8.8
dre:100330633 tripartite motif-containing protein 3-like 28.1 9.6
> tgo:TGME49_064450 DNA topoisomerase III beta-1, putative (EC:5.99.1.2);
K03165 DNA topoisomerase III [EC:5.99.1.2]
Length=841
Score = 182 bits (463), Expect = 2e-46, Method: Compositional matrix adjust.
Identities = 80/141 (56%), Positives = 104/141 (73%), Gaps = 1/141 (0%)
Query 1 TIKLYKELKCPLDGFELLLSVSVGGKAFSFCPLCYNDPPFDMKGQKTMSCVDCLHPSCRH 60
IKLYKEL+CPLD FEL+L GGK+F CPLCYNDPP + K MSCV+C HP+C+H
Sbjct 639 AIKLYKELRCPLDNFELVLFTQKGGKSFPLCPLCYNDPPLEGSVTK-MSCVECTHPTCKH 697
Query 61 SINAIGVMRCPQENCQGLLYLNPVSAPRWRLDCTFCRYELLLFEGAFKVAVTDDECSSCG 120
I A+GV++CPQ +C+G+LYL+ VS P+W+LDC C+Y++ L EGA K++VT DEC +CG
Sbjct 698 GIEAVGVLQCPQPDCKGILYLDVVSGPKWKLDCNACQYQVKLMEGAHKISVTPDECDTCG 757
Query 121 SMQLEVTFSKANPLKDGSTER 141
S L V +K P KDG E+
Sbjct 758 SALLNVVMNKVKPFKDGEVEK 778
> dre:100329289 topoisomerase (DNA) III beta-like; K03165 DNA
topoisomerase III [EC:5.99.1.2]
Length=861
Score = 129 bits (324), Expect = 3e-30, Method: Compositional matrix adjust.
Identities = 69/144 (47%), Positives = 95/144 (65%), Gaps = 8/144 (5%)
Query 2 IKLYKELKCPLDGFELLLSVSVG-GKAFSFCPLCYNDPPF-DMKGQKTMSCVDCLHPSCR 59
IKLYKELKCPLD FEL+L S GK++ CP C+++PPF DMK K M C +C HPSC+
Sbjct 658 IKLYKELKCPLDEFELVLWTSGSRGKSYPVCPYCFSNPPFRDMK--KGMGCNECTHPSCQ 715
Query 60 HSINAIGVMRCPQENCQ-GLLYLNPVSAPRWRLDCTFCRYELLLFEGAFKVAVTDDECSS 118
HS+N++G+ +C + C+ G+L +P S P+WR+ C C + FE A KV V+ D C S
Sbjct 716 HSLNSLGIGQCVE--CETGVLVFDPTSGPKWRMACNKCNVVVHFFEQAHKVQVSQDSCDS 773
Query 119 CGSMQLEVTFSKA-NPLKDGSTER 141
C + + V F+K +PL DG T+
Sbjct 774 CEASLVIVDFNKTRSPLPDGETQH 797
> mmu:21976 Top3b; topoisomerase (DNA) III beta (EC:5.99.1.2);
K03165 DNA topoisomerase III [EC:5.99.1.2]
Length=862
Score = 125 bits (315), Expect = 3e-29, Method: Compositional matrix adjust.
Identities = 68/145 (46%), Positives = 95/145 (65%), Gaps = 8/145 (5%)
Query 1 TIKLYKELKCPLDGFELLL-SVSVGGKAFSFCPLCYNDPPF-DMKGQKTMSCVDCLHPSC 58
TIKLYKEL+CPLD FEL+L S GK++ CP CYN PPF DMK K M C +C HP+C
Sbjct 657 TIKLYKELRCPLDDFELVLWSSGSRGKSYPLCPYCYNHPPFRDMK--KGMGCNECTHPTC 714
Query 59 RHSINAIGVMRCPQ-ENCQGLLYLNPVSAPRWRLDCTFCRYELLLFEGAFKVAVTDDECS 117
+HS++ +G+ +C + EN G+L L+P S P+W++ C C FE A +V V+ D C+
Sbjct 715 QHSLSMLGIGQCVECEN--GVLVLDPTSGPKWKVACNTCNVVAHCFENAHRVRVSADTCN 772
Query 118 SCGSMQLEVTFSKA-NPLKDGSTER 141
+C + L+V F+KA +PL T+
Sbjct 773 TCEAALLDVDFNKAKSPLPGNETQH 797
> hsa:8940 TOP3B, FLJ39376; topoisomerase (DNA) III beta (EC:5.99.1.2);
K03165 DNA topoisomerase III [EC:5.99.1.2]
Length=862
Score = 125 bits (313), Expect = 6e-29, Method: Compositional matrix adjust.
Identities = 69/145 (47%), Positives = 94/145 (64%), Gaps = 8/145 (5%)
Query 1 TIKLYKELKCPLDGFELLL-SVSVGGKAFSFCPLCYNDPPF-DMKGQKTMSCVDCLHPSC 58
TIKLYKEL+CPLD FEL+L S GK++ CP CYN PPF DMK K M C +C HPSC
Sbjct 657 TIKLYKELRCPLDDFELVLWSSGSRGKSYPLCPYCYNHPPFRDMK--KGMGCNECTHPSC 714
Query 59 RHSINAIGVMRCPQENCQ-GLLYLNPVSAPRWRLDCTFCRYELLLFEGAFKVAVTDDECS 117
+HS++ +G+ +C + C+ G+L L+P S P+W++ C C FE A +V V+ D CS
Sbjct 715 QHSLSMLGIGQCVE--CESGVLVLDPTSGPKWKVACNKCNVVAHCFENAHRVRVSADTCS 772
Query 118 SCGSMQLEVTFSKA-NPLKDGSTER 141
C + L+V F+KA +PL T+
Sbjct 773 VCEAALLDVDFNKAKSPLPGDETQH 797
> ath:AT2G32000 DNA topoisomerase family protein; K03165 DNA topoisomerase
III [EC:5.99.1.2]
Length=838
Score = 124 bits (311), Expect = 9e-29, Method: Compositional matrix adjust.
Identities = 67/159 (42%), Positives = 91/159 (57%), Gaps = 22/159 (13%)
Query 1 TIKLYKELKCPLDGFELLLSVSVG--GKAFSFCPLCYNDPPFD----------------- 41
T+KLYKEL CPLD FEL++ G GK+F CP CYN PPF+
Sbjct 624 TVKLYKELTCPLDNFELVIYSVPGPEGKSFPLCPYCYNSPPFEGIDTLFGASKTPNAPAK 683
Query 42 MKGQKTMSCVDCLHPSCRHSINAIGVMRCPQENCQGLLYLNPVSAPRWRLDCTFCRYELL 101
K M C C HP+C+HS+ GV CP+ C+G L L+PVS P+W+L+C C +L
Sbjct 684 TKTGAGMPCSLCPHPTCQHSVRNQGVCACPE--CEGTLVLDPVSFPKWKLNCNLCSCIVL 741
Query 102 LFEGAFKVAVTDDECSSCGSMQLEVTFS-KANPLKDGST 139
L EGA ++ T + C C S +E+ F+ K PL++G+T
Sbjct 742 LPEGAHRITTTSNRCPECDSAIIEIDFNKKTTPLENGAT 780
> xla:398564 top3b, MGC53016; topoisomerase (DNA) III beta (EC:5.99.1.2);
K03165 DNA topoisomerase III [EC:5.99.1.2]
Length=858
Score = 117 bits (293), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 62/145 (42%), Positives = 92/145 (63%), Gaps = 8/145 (5%)
Query 1 TIKLYKELKCPLDGFELLL-SVSVGGKAFSFCPLCYNDPPF-DMKGQKTMSCVDCLHPSC 58
TIKLYKEL+CPLD FEL+L S GK++ CP CYN PF DMK K M C +C HP+C
Sbjct 657 TIKLYKELRCPLDDFELVLWSSGSRGKSYPLCPYCYNHTPFRDMK--KGMGCNECTHPTC 714
Query 59 RHSINAIGVMRCPQENCQ-GLLYLNPVSAPRWRLDCTFCRYELLLFEGAFKVAVTDDECS 117
+HS++ +G+ +C + C+ G++ L+ S P+W++ C C + FE A +V V+ + C
Sbjct 715 QHSLSMLGIAQCVE--CESGVMVLDSTSGPKWKMACNKCNVIVHFFENAHRVRVSAETCE 772
Query 118 SCGSMQLEVTFSKA-NPLKDGSTER 141
+C + + V F+KA +PL T+
Sbjct 773 ACDAALVHVDFNKAKSPLPADQTQH 797
> bbo:BBOV_II000360 18.m06014; DNA topoisomerase; K03165 DNA topoisomerase
III [EC:5.99.1.2]
Length=878
Score = 114 bits (285), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 67/153 (43%), Positives = 88/153 (57%), Gaps = 24/153 (15%)
Query 1 TIKLYKELKCPLDGFELLLSVSVGGKAFSFCPLCYNDPPFDMKGQKTMSCVDCLHPSCRH 60
TIKLYK +KCPLD FELL +AFS CP+CYNDPPF+ G K M C+DC HP+C +
Sbjct 681 TIKLYKGIKCPLDNFELL-QFQSRLRAFSLCPMCYNDPPFESIGNKGMGCIDCAHPTCAN 739
Query 61 SINAIGVMRCPQENCQGLLYLNPVS-APRWRLDCTFCRYE--------------LLLFEG 105
S +A+ + RCP + C G+L L+ ++ AP W+ C+ C Y+ LL +
Sbjct 740 SADALTLTRCPGD-CDGMLILDIINGAPNWKFFCSSCSYKLQFSDITKSRFDRLLLDYHE 798
Query 106 AFKVA-VTDDECSSCGSMQLEVTFSKANPLKDG 137
+VA +T EC SC S EV LKDG
Sbjct 799 RIEVAIITGKECDSCESQLTEVI------LKDG 825
> cpv:cgd6_2390 DNA topoisomerase III beta-1 ; K03165 DNA topoisomerase
III [EC:5.99.1.2]
Length=835
Score = 102 bits (254), Expect = 4e-22, Method: Compositional matrix adjust.
Identities = 50/127 (39%), Positives = 74/127 (58%), Gaps = 2/127 (1%)
Query 1 TIKLYKELKCPLDGFELLLSVSVGGKAFSFCPLCYNDPPFDMKGQKTMSCVDCLHPSCRH 60
TIK+YKELKCP+D FELLL + GK FCP CYNDPPF + ++ M C C H +C
Sbjct 657 TIKIYKELKCPIDDFELLLFIDQKGKKSIFCPRCYNDPPF-LDAKENMLCKFCPHQTCNF 715
Query 61 SINAIGVMRCPQENCQ-GLLYLNPVSAPRWRLDCTFCRYELLLFEGAFKVAVTDDECSSC 119
S+ + M CP ++C+ G++ ++ S WR DC+ C + + + + C C
Sbjct 716 SLKSTFFMACPSQDCRDGIITMDSSSTSDWRFDCSRCSISFSIKKNICERLSLGERCEKC 775
Query 120 GSMQLEV 126
GS +L++
Sbjct 776 GSRKLKI 782
> cel:Y48C3A.14 hypothetical protein; K03165 DNA topoisomerase
III [EC:5.99.1.2]
Length=855
Score = 99.8 bits (247), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 57/143 (39%), Positives = 85/143 (59%), Gaps = 7/143 (4%)
Query 2 IKLYKELKCPLDGFELLLSVSVGGK---AFSFCPLCYNDPPFDMKGQKTMSCVDCLHPSC 58
+K+ + KCPLD F+++L GGK F+ CP C+N+PPF+ K + C C+HPSC
Sbjct 649 LKVIGDHKCPLDDFDVILYQGPGGKLARTFTLCPFCFNNPPFE-KMPQGAGCDTCVHPSC 707
Query 59 RHSINAIGVMRCPQENCQGLLYLNPVSAPRWRLDCTFCRYELLLFEGAFKVAV-TDDECS 117
++SI++ GV C Q+ C G++ L+ S P+WR+ C C + LFEGA KV V + C
Sbjct 708 QYSISSNGVCGCLQD-CGGVMLLDTQSHPKWRMTCNKCPSVVGLFEGAIKVKVDSTRSCE 766
Query 118 SCGSMQLEVTFSKANPLKDGSTE 140
C + ++ F K N K G+ E
Sbjct 767 QCDAQFIKAEF-KPNSGKPGAIE 788
> ath:AT3G45510 zinc finger (C3HC4-type RING finger) family protein
Length=257
Score = 33.1 bits (74), Expect = 0.26, Method: Compositional matrix adjust.
Identities = 20/60 (33%), Positives = 30/60 (50%), Gaps = 11/60 (18%)
Query 31 CPLCYNDPPFDMKGQKTMS--------CVDCLHPSCRHSINAIGVMRCPQENCQGLLYLN 82
C +C+ D D++GQ+ S CV+C+ S+N GV RCP+ C+ L L
Sbjct 66 CGICFVD---DIEGQEMFSAALCSHYFCVECMKQRIEVSLNEGGVPRCPRHGCKSALTLR 122
> hsa:54542 RC3H2, FLJ20301, FLJ20713, MGC52176, MNAB, RNF164;
ring finger and CCCH-type domains 2
Length=1191
Score = 32.7 bits (73), Expect = 0.39, Method: Composition-based stats.
Identities = 16/44 (36%), Positives = 23/44 (52%), Gaps = 3/44 (6%)
Query 28 FSFCPLCYNDPPFDMKGQKTMSCVDCLHPSCRHSINAIGVMRCP 71
F CP+CYN+ FD K +S + C H C+ +N + CP
Sbjct 11 FLSCPICYNE--FDENVHKPIS-LGCSHTVCKTCLNKLHRKACP 51
> mmu:319817 Rc3h2, 2900024N03Rik, 9430019J22Rik, D930043C02Rik,
Mnab, Rnf164; ring finger and CCCH-type zinc finger domains
2
Length=1187
Score = 32.7 bits (73), Expect = 0.40, Method: Composition-based stats.
Identities = 16/44 (36%), Positives = 23/44 (52%), Gaps = 3/44 (6%)
Query 28 FSFCPLCYNDPPFDMKGQKTMSCVDCLHPSCRHSINAIGVMRCP 71
F CP+CYN+ FD K +S + C H C+ +N + CP
Sbjct 11 FLSCPICYNE--FDENVHKPIS-LGCSHTVCKTCLNKLHRKACP 51
> cel:F56D2.5 hypothetical protein; K11971 E3 ubiquitin-protein
ligase RNF14 [EC:6.3.2.19]
Length=437
Score = 30.8 bits (68), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 13/31 (41%), Positives = 19/31 (61%), Gaps = 2/31 (6%)
Query 68 MRCPQENCQGLLYLNPVSAPRWRLDCTFCRY 98
M CP ENCQ + YL + R ++C++C Y
Sbjct 271 MECPNENCQMVAYL--TDSQRNLVECSYCNY 299
> ath:AT3G45480 zinc ion binding
Length=382
Score = 30.4 bits (67), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 21/73 (28%), Positives = 33/73 (45%), Gaps = 11/73 (15%)
Query 17 LLLSVSVGGKAFSFCPLCYNDPPFDMKGQKTMS--------CVDCLHPSCRHSINAIGVM 68
L+ ++S+ C C+ND +KG+ S CV+C+ S+N G+
Sbjct 142 LVSNISIPRPQKKTCGNCFND---GIKGENMFSADLCSHYFCVECMKEHIEVSLNEGGLP 198
Query 69 RCPQENCQGLLYL 81
RCP + C L L
Sbjct 199 RCPHDGCTSNLTL 211
> tgo:TGME49_074140 RNA recognition motif-containing protein
Length=788
Score = 29.6 bits (65), Expect = 3.3, Method: Composition-based stats.
Identities = 13/48 (27%), Positives = 23/48 (47%), Gaps = 0/48 (0%)
Query 75 CQGLLYLNPVSAPRWRLDCTFCRYELLLFEGAFKVAVTDDECSSCGSM 122
C+ + +PV P W L C+ C ++ L ++ +C CGS+
Sbjct 716 CRKFIAFDPVLLPCWHLCCSDCVHQCLRWQPNHPSEPVVVDCPECGSV 763
> xla:100137626 tnfrsf1a, cd120a, fpf, p55, p55-r, tbp1, tnf-r,
tnfar, tnfr1, tnfr55, tnfr60, xtnfr1; tumor necrosis factor
receptor superfamily, member 1A; K03158 tumor necrosis factor
receptor superfamily member 1A
Length=412
Score = 28.5 bits (62), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 21/57 (36%), Positives = 29/57 (50%), Gaps = 2/57 (3%)
Query 19 LSVSVGGKAFSFCPLCYNDPPFDMKGQKTMSCVDCLHPSCR-HSINAIGVMRCPQEN 74
+SV G + C C+ FD +K +SC C P+CR H A G++ PQEN
Sbjct 166 VSVPCNGYNDTIC-TCHFGFYFDRGSRKCLSCDICNDPTCRQHCPPASGLVNPPQEN 221
> ath:AT5G53740 hypothetical protein
Length=275
Score = 28.1 bits (61), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 28/104 (26%), Positives = 39/104 (37%), Gaps = 3/104 (2%)
Query 2 IKLYKELKCPLDGFELLLSVSVGGKAFSFCPLCYNDPPFDMKGQKTMSCVDCLHPSCRHS 61
I L K + P E + S K S CP C PPF + TM + C +
Sbjct 71 INLSKNRRMPFKSRENTIFFSKRRKNSSLCPHC-TAPPFQLS--PTMLLMFCHDGARLKG 127
Query 62 INAIGVMRCPQENCQGLLYLNPVSAPRWRLDCTFCRYELLLFEG 105
+N +GL+ +S P +D T C LL +G
Sbjct 128 MNPRNAEERKYRQAEGLVTPQFLSIPGSPIDLTKCWSSLLNIQG 171
> dre:100330633 tripartite motif-containing protein 3-like
Length=281
Score = 28.1 bits (61), Expect = 9.6, Method: Compositional matrix adjust.
Identities = 21/77 (27%), Positives = 36/77 (46%), Gaps = 23/77 (29%)
Query 26 KAFSF----CPLCYNDPPFDMKGQKTMSCVDCLHPSCRHSINAIGVMRCPQENCQGLLYL 81
KAF + C +CYN FD+ G++ ++CLH C ++A+ + C ++
Sbjct 45 KAFPYEEYECKICYNY--FDL-GRRAPKILECLHTFCEECLHALQL--CEEQP------- 92
Query 82 NPVSAPRWRLDCTFCRY 98
WR+ C CR+
Sbjct 93 -------WRISCPVCRH 102
Lambda K H
0.324 0.139 0.458
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2618291680
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40