bitscore colors: <40, 40-50 , 50-80, 80-200, >200
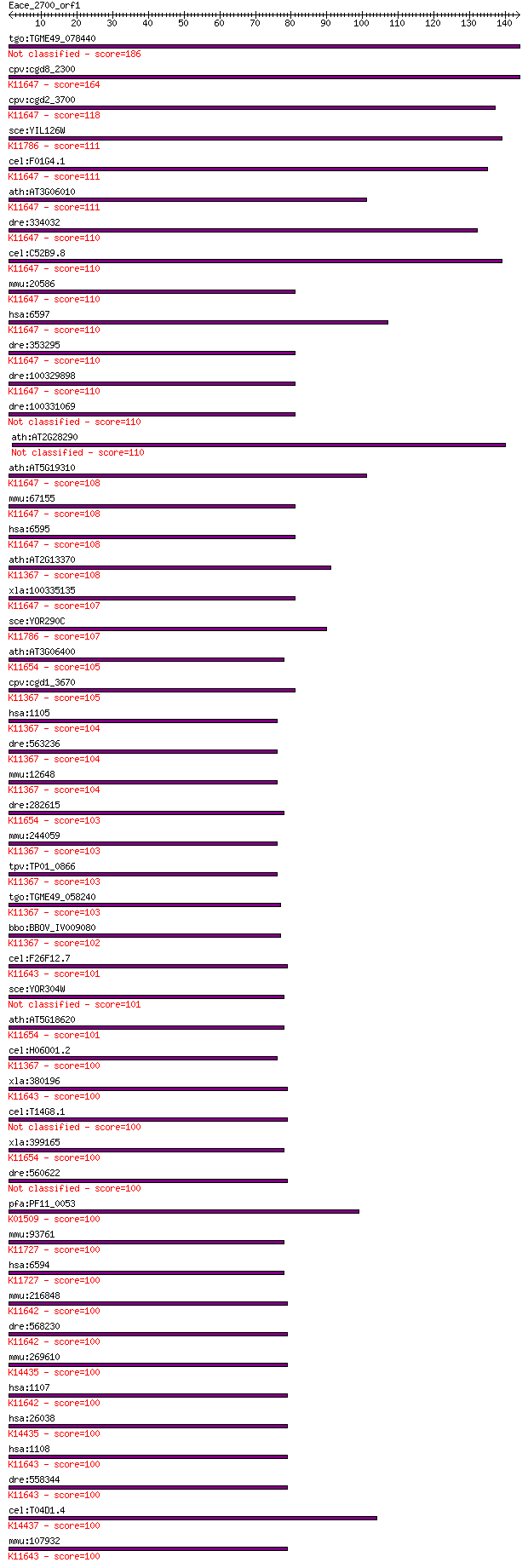
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2700_orf1
Length=143
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_078440 transcription regulatory protein SNF2, putat... 186 2e-47
cpv:cgd8_2300 brahma like protein with a HSA domain, SNF2 like... 164 7e-41
cpv:cgd2_3700 SWI/SNF related transcriptional regulator ATpase... 118 5e-27
sce:YIL126W STH1, NPS1; ATPase component of the RSC chromatin ... 111 7e-25
cel:F01G4.1 psa-4; Phasmid Socket Absent family member (psa-4)... 111 8e-25
ath:AT3G06010 ATCHR12; ATP binding / DNA binding / helicase/ n... 111 9e-25
dre:334032 smarca2, psa-4, wu:fa56c07, wu:fi27f11, zgc:66238; ... 110 1e-24
cel:C52B9.8 hypothetical protein; K11647 SWI/SNF-related matri... 110 1e-24
mmu:20586 Smarca4, Brg1, HP1-BP72, SNF2beta, SW1/SNF; SWI/SNF ... 110 1e-24
hsa:6597 SMARCA4, BAF190, BRG1, FLJ39786, RTPS2, SNF2, SNF2-BE... 110 1e-24
dre:353295 smarca4, brg1, smarca2, wu:fb51a08; SWI/SNF related... 110 2e-24
dre:100329898 SWI/SNF-related matrix-associated actin-dependen... 110 2e-24
dre:100331069 SWI/SNF-related matrix-associated actin-dependen... 110 2e-24
ath:AT2G28290 SYD; SYD (SPLAYED); ATPase/ chromatin binding 110 2e-24
ath:AT5G19310 homeotic gene regulator, putative; K11647 SWI/SN... 108 5e-24
mmu:67155 Smarca2, 2610209L14Rik, SNF2alpha, Snf2l2, brahma, b... 108 5e-24
hsa:6595 SMARCA2, BAF190, BRM, FLJ36757, MGC74511, SNF2, SNF2L... 108 5e-24
ath:AT2G13370 CHR5; CHR5 (chromatin remodeling 5); ATP binding... 108 8e-24
xla:100335135 smarca4, baf190, brg1, rtps2, snf2, snf2-beta, s... 107 9e-24
sce:YOR290C SNF2, GAM1, HAF1, SWI2, TYE3; Catalytic subunit of... 107 1e-23
ath:AT3G06400 CHR11; CHR11 (CHROMATIN-REMODELING PROTEIN 11); ... 105 5e-23
cpv:cgd1_3670 CHD3 ortholog with 2x chromodomains plus SNF2 AT... 105 7e-23
hsa:1105 CHD1, DKFZp686E2337; chromodomain helicase DNA bindin... 104 8e-23
dre:563236 MGC173506, wu:fc26h11, wu:fk85d05; zgc:173506; K113... 104 8e-23
mmu:12648 Chd1, 4930525N21Rik, AI851787, AW555109, MGC141554; ... 104 8e-23
dre:282615 smarca5, chunp6878, fb26d12, fb49g04, im:7146484, w... 103 2e-22
mmu:244059 Chd2, 2810013C04Rik, 2810040A01Rik, 5630401D06Rik, ... 103 2e-22
tpv:TP01_0866 hypothetical protein; K11367 chromodomain-helica... 103 2e-22
tgo:TGME49_058240 chromodomain helicase DNA binding protein, p... 103 2e-22
bbo:BBOV_IV009080 23.m05963; chromo-helicase DNA-binding prote... 102 6e-22
cel:F26F12.7 let-418; LEThal family member (let-418); K11643 c... 101 6e-22
sce:YOR304W ISW2; ATP-dependent DNA translocase involved in ch... 101 6e-22
ath:AT5G18620 CHR17; CHR17 (CHROMATIN REMODELING FACTOR17); AT... 101 8e-22
cel:H06O01.2 hypothetical protein; K11367 chromodomain-helicas... 100 1e-21
xla:380196 chd4, MGC52739, b230399n07; chromodomain helicase D... 100 1e-21
cel:T14G8.1 chd-3; CHromoDomain protein family member (chd-3) 100 1e-21
xla:399165 smarca5, iswi; SWI/SNF related, matrix associated, ... 100 1e-21
dre:560622 fd12d03, wu:fb44b12; wu:fd12d03 100 1e-21
pfa:PF11_0053 PfSNF2L; K01509 adenosinetriphosphatase [EC:3.6.... 100 1e-21
mmu:93761 Smarca1, 5730494M04Rik, Snf2l; SWI/SNF related, matr... 100 1e-21
hsa:6594 SMARCA1, DKFZp686D1623, FLJ41547, ISWI, NURF140, SNF2... 100 2e-21
mmu:216848 Chd3, 2600010P09Rik, AF020312, Chd7, MGC40857, Prp7... 100 2e-21
dre:568230 similar to CHD3; K11642 chromodomain-helicase-DNA-b... 100 2e-21
mmu:269610 Chd5, 4930532L22Rik, AW060752, B230399N07Rik; chrom... 100 2e-21
hsa:1107 CHD3, Mi-2a, Mi2-ALPHA, ZFH; chromodomain helicase DN... 100 2e-21
hsa:26038 CHD5, DKFZp434N231, KIAA0444; chromodomain helicase ... 100 2e-21
hsa:1108 CHD4, DKFZp686E06161, Mi-2b, Mi2-BETA; chromodomain h... 100 2e-21
dre:558344 im:7143343; si:ch211-51m24.3; K11643 chromodomain-h... 100 2e-21
cel:T04D1.4 tag-192; Temporarily Assigned Gene name family mem... 100 2e-21
mmu:107932 Chd4, 9530019N15Rik, AA617397, BC005710, D6Ertd380e... 100 2e-21
> tgo:TGME49_078440 transcription regulatory protein SNF2, putative
(EC:2.7.11.1 3.4.21.97 3.2.1.3 3.1.3.33 3.4.24.61 3.4.24.35)
Length=2668
Score = 186 bits (472), Expect = 2e-47, Method: Compositional matrix adjust.
Identities = 95/149 (63%), Positives = 116/149 (77%), Gaps = 6/149 (4%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQK+EVLT+RF++ SIEEQIL
Sbjct 1650 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKKEVLTLRFISVESIEEQIL 1709
Query 61 HSAELKLDKDALVIKSGMYNGELQD------RETERQEQVREILRRRKQLEANWSRPFDF 114
AE KLDKD LVI+SGMY G Q+ R+ ER QVREILR+++QL+ N +R D
Sbjct 1710 QRAECKLDKDKLVIQSGMYYGHGQEEVHDPSRDLERTNQVREILRKQRQLDVNLTRALDL 1769
Query 115 GILKSQLARNPKEATLFDALQKIRQLLRL 143
+LK Q+AR+ ++ +F+ IR+LL +
Sbjct 1770 QLLKRQIARSSEDMRVFERADCIRRLLHI 1798
> cpv:cgd8_2300 brahma like protein with a HSA domain, SNF2 like
helicase and a bromo domain ; K11647 SWI/SNF-related matrix-associated
actin-dependent regulator of chromatin subfamily
A member 2/4 [EC:3.6.4.-]
Length=1673
Score = 164 bits (416), Expect = 7e-41, Method: Compositional matrix adjust.
Identities = 81/145 (55%), Positives = 115/145 (79%), Gaps = 3/145 (2%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
TKAGG G+NLQSADTVI+FDSDWNPQNDEQAQSRAHRIGQK+EVLT+RFVTP ++EE+I+
Sbjct 1135 TKAGGFGINLQSADTVILFDSDWNPQNDEQAQSRAHRIGQKKEVLTLRFVTPDTVEERIM 1194
Query 61 HSAELKLDKDALVIKSGMYNG--ELQDRETERQEQVREILRRRKQLEANWSRPFDFGILK 118
+A +KLDKDAL+IKSGMY+ + D E +R+E+++EILR+++Q E + +D L
Sbjct 1195 TTAGIKLDKDALIIKSGMYHDLYDGDDLEQKRKEKIQEILRKQRQKEV-VNCYYDSDRLN 1253
Query 119 SQLARNPKEATLFDALQKIRQLLRL 143
LAR+ ++ +F+ + ++R++ +
Sbjct 1254 RILARSDRDLEIFERVDRMRKMCHI 1278
> cpv:cgd2_3700 SWI/SNF related transcriptional regulator ATpase
; K11647 SWI/SNF-related matrix-associated actin-dependent
regulator of chromatin subfamily A member 2/4 [EC:3.6.4.-]
Length=1552
Score = 118 bits (296), Expect = 5e-27, Method: Compositional matrix adjust.
Identities = 64/137 (46%), Positives = 93/137 (67%), Gaps = 5/137 (3%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
T+AGGLG+NLQ+ADTVIIFDSDWNP D QAQSRAHR+GQK EV +RFV+ S +EE +L
Sbjct 966 TRAGGLGLNLQAADTVIIFDSDWNPHQDLQAQSRAHRMGQKNEVRVLRFVSISGVEELVL 1025
Query 61 HSAELKLDKDALVIKSGMYNGELQDRETERQEQVREIL-RRRKQLEANWSRPFDFGILKS 119
A+ KL+ D +I++GM+N Q E ER+++++E+ + + ++ + P +
Sbjct 1026 KRAQKKLEIDHKIIQAGMFNS-TQVEEEEREDRLKELFGKEEYKSDSRVTTPSEINQF-- 1082
Query 120 QLARNPKEATLFDALQK 136
LARN +E F+ + K
Sbjct 1083 -LARNDEELKAFEEMDK 1098
> sce:YIL126W STH1, NPS1; ATPase component of the RSC chromatin
remodeling complex; required for expression of early meiotic
genes; essential helicase-related protein homologous to Snf2p
(EC:3.6.1.-); K11786 ATP-dependent helicase STH1/SNF2 [EC:3.6.4.-]
Length=1359
Score = 111 bits (278), Expect = 7e-25, Method: Compositional matrix adjust.
Identities = 68/142 (47%), Positives = 85/142 (59%), Gaps = 10/142 (7%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
T+AGGLG+NLQ+ADTVIIFD+DWNP D QAQ RAHRIGQK EV +R +T S+EE IL
Sbjct 867 TRAGGLGLNLQTADTVIIFDTDWNPHQDLQAQDRAHRIGQKNEVRILRLITTDSVEEVIL 926
Query 61 HSAELKLDKDALVIKSGMYNGELQDRETERQEQVREILRRRKQLEANWSRPFDFGI---- 116
A KLD D VI++G + D ++ +EQ LRR + E N +
Sbjct 927 ERAMQKLDIDGKVIQAGKF-----DNKSTAEEQ-EAFLRRLIESETNRDDDDKAELDDDE 980
Query 117 LKSQLARNPKEATLFDALQKIR 138
L LAR+ E LFD + K R
Sbjct 981 LNDTLARSADEKILFDKIDKER 1002
> cel:F01G4.1 psa-4; Phasmid Socket Absent family member (psa-4);
K11647 SWI/SNF-related matrix-associated actin-dependent
regulator of chromatin subfamily A member 2/4 [EC:3.6.4.-]
Length=1474
Score = 111 bits (277), Expect = 8e-25, Method: Compositional matrix adjust.
Identities = 62/136 (45%), Positives = 91/136 (66%), Gaps = 7/136 (5%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
T+AGGLG+NLQ+ADTVIIFDSDWNP D QAQ RAHRIGQK+EV +R +T +S+EE+IL
Sbjct 926 TRAGGLGLNLQTADTVIIFDSDWNPHQDMQAQDRAHRIGQKKEVRVLRLITANSVEEKIL 985
Query 61 HSAELKLDKDALVIKSGMYNGELQDRET--ERQEQVREILRRRKQLEANWSRPFDFGILK 118
+A KL+ D VI++G ++ R T ER++ + +I++ + E P D +
Sbjct 986 AAARYKLNVDEKVIQAGKFD----QRSTGAERKQMLEQIIQADGEEEEEEEVPDDETV-N 1040
Query 119 SQLARNPKEATLFDAL 134
+AR+ +E +F ++
Sbjct 1041 QMVARSEEEFNIFQSM 1056
> ath:AT3G06010 ATCHR12; ATP binding / DNA binding / helicase/
nucleic acid binding; K11647 SWI/SNF-related matrix-associated
actin-dependent regulator of chromatin subfamily A member
2/4 [EC:3.6.4.-]
Length=1132
Score = 111 bits (277), Expect = 9e-25, Method: Compositional matrix adjust.
Identities = 55/100 (55%), Positives = 74/100 (74%), Gaps = 2/100 (2%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
T+AGGLG+NLQ+ADTVIIFDSDWNPQ D+QA+ RAHRIGQK+EV V+ S+EE IL
Sbjct 819 TRAGGLGLNLQTADTVIIFDSDWNPQMDQQAEDRAHRIGQKKEVRVFVLVSVGSVEEVIL 878
Query 61 HSAELKLDKDALVIKSGMYNGELQDRETERQEQVREILRR 100
A+ K+ DA VI++G++N +R+E + EI+R+
Sbjct 879 ERAKQKMGIDAKVIQAGLFN--TTSTAQDRREMLEEIMRK 916
> dre:334032 smarca2, psa-4, wu:fa56c07, wu:fi27f11, zgc:66238;
SWI/SNF related, matrix associated, actin dependent regulator
of chromatin, subfamily a, member 2; K11647 SWI/SNF-related
matrix-associated actin-dependent regulator of chromatin
subfamily A member 2/4 [EC:3.6.4.-]
Length=1568
Score = 110 bits (276), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 63/131 (48%), Positives = 85/131 (64%), Gaps = 3/131 (2%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
T+AGGLG+NLQ+ADTV+IFDSDWNP D QAQ RAHRIGQ+ EV +R T +S+EE+IL
Sbjct 1128 TRAGGLGLNLQAADTVVIFDSDWNPHQDLQAQDRAHRIGQQNEVRVLRLCTVNSVEEKIL 1187
Query 61 HSAELKLDKDALVIKSGMYNGELQDRETERQEQVREILRRRKQLEANWSRPFDFGILKSQ 120
+A+ KL+ D VI++GM++ + ER+ ++ IL +Q P D L
Sbjct 1188 AAAKYKLNVDQKVIQAGMFDQ--KSSSHERRAFLQAILEHEEQNMEEDEVP-DDETLNQM 1244
Query 121 LARNPKEATLF 131
+ARN E LF
Sbjct 1245 IARNEDEFELF 1255
> cel:C52B9.8 hypothetical protein; K11647 SWI/SNF-related matrix-associated
actin-dependent regulator of chromatin subfamily
A member 2/4 [EC:3.6.4.-]
Length=1336
Score = 110 bits (276), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 64/138 (46%), Positives = 89/138 (64%), Gaps = 3/138 (2%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
T+AGGLG+NLQ+ADTVIIFDSDWNP D QAQ RAHRIGQK EV R +T +S+EE+IL
Sbjct 759 TRAGGLGLNLQTADTVIIFDSDWNPHQDMQAQDRAHRIGQKAEVRVFRLITANSVEEKIL 818
Query 61 HSAELKLDKDALVIKSGMYNGELQDRETERQEQVREILRRRKQLEANWSRPFDFGILKSQ 120
+A KL+ D VI++G ++ + ER+E + I++ + E + P D I
Sbjct 819 AAARYKLNVDEKVIQAGKFDN--RSTGAERREILENIIKTENESEEDEEVPNDEDI-NDI 875
Query 121 LARNPKEATLFDALQKIR 138
L+R+ +E LF + + R
Sbjct 876 LSRSEEEFELFQKMDQER 893
> mmu:20586 Smarca4, Brg1, HP1-BP72, SNF2beta, SW1/SNF; SWI/SNF
related, matrix associated, actin dependent regulator of chromatin,
subfamily a, member 4; K11647 SWI/SNF-related matrix-associated
actin-dependent regulator of chromatin subfamily
A member 2/4 [EC:3.6.4.-]
Length=1617
Score = 110 bits (275), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 51/80 (63%), Positives = 65/80 (81%), Gaps = 0/80 (0%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
T+AGGLG+NLQSADTVIIFDSDWNP D QAQ RAHRIGQ+ EV +R T +S+EE+IL
Sbjct 1156 TRAGGLGLNLQSADTVIIFDSDWNPHQDLQAQDRAHRIGQQNEVRVLRLCTVNSVEEKIL 1215
Query 61 HSAELKLDKDALVIKSGMYN 80
+A+ KL+ D VI++GM++
Sbjct 1216 AAAKYKLNVDQKVIQAGMFD 1235
> hsa:6597 SMARCA4, BAF190, BRG1, FLJ39786, RTPS2, SNF2, SNF2-BETA,
SNF2L4, SNF2LB, SWI2, hSNF2b; SWI/SNF related, matrix
associated, actin dependent regulator of chromatin, subfamily
a, member 4; K11647 SWI/SNF-related matrix-associated actin-dependent
regulator of chromatin subfamily A member 2/4 [EC:3.6.4.-]
Length=1647
Score = 110 bits (275), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 57/106 (53%), Positives = 77/106 (72%), Gaps = 2/106 (1%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
T+AGGLG+NLQSADTVIIFDSDWNP D QAQ RAHRIGQ+ EV +R T +S+EE+IL
Sbjct 1156 TRAGGLGLNLQSADTVIIFDSDWNPHQDLQAQDRAHRIGQQNEVRVLRLCTVNSVEEKIL 1215
Query 61 HSAELKLDKDALVIKSGMYNGELQDRETERQEQVREILRRRKQLEA 106
+A+ KL+ D VI++GM++ + ER+ ++ IL +Q E+
Sbjct 1216 AAAKYKLNVDQKVIQAGMFDQ--KSSSHERRAFLQAILEHEEQDES 1259
> dre:353295 smarca4, brg1, smarca2, wu:fb51a08; SWI/SNF related,
matrix associated, actin dependent regulator of chromatin,
subfamily a, member 4; K11647 SWI/SNF-related matrix-associated
actin-dependent regulator of chromatin subfamily A member
2/4 [EC:3.6.4.-]
Length=1627
Score = 110 bits (275), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 51/80 (63%), Positives = 65/80 (81%), Gaps = 0/80 (0%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
T+AGGLG+NLQSADTVIIFDSDWNP D QAQ RAHRIGQ+ EV +R T +S+EE+IL
Sbjct 1168 TRAGGLGLNLQSADTVIIFDSDWNPHQDLQAQDRAHRIGQQNEVRVLRLCTVNSVEEKIL 1227
Query 61 HSAELKLDKDALVIKSGMYN 80
+A+ KL+ D VI++GM++
Sbjct 1228 AAAKYKLNVDQKVIQAGMFD 1247
> dre:100329898 SWI/SNF-related matrix-associated actin-dependent
regulator of chromatin a4-like; K11647 SWI/SNF-related matrix-associated
actin-dependent regulator of chromatin subfamily
A member 2/4 [EC:3.6.4.-]
Length=1627
Score = 110 bits (275), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 51/80 (63%), Positives = 65/80 (81%), Gaps = 0/80 (0%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
T+AGGLG+NLQSADTVIIFDSDWNP D QAQ RAHRIGQ+ EV +R T +S+EE+IL
Sbjct 1168 TRAGGLGLNLQSADTVIIFDSDWNPHQDLQAQDRAHRIGQQNEVRVLRLCTVNSVEEKIL 1227
Query 61 HSAELKLDKDALVIKSGMYN 80
+A+ KL+ D VI++GM++
Sbjct 1228 AAAKYKLNVDQKVIQAGMFD 1247
> dre:100331069 SWI/SNF-related matrix-associated actin-dependent
regulator of chromatin a4-like
Length=1234
Score = 110 bits (274), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 51/80 (63%), Positives = 65/80 (81%), Gaps = 0/80 (0%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
T+AGGLG+NLQSADTVIIFDSDWNP D QAQ RAHRIGQ+ EV +R T +S+EE+IL
Sbjct 775 TRAGGLGLNLQSADTVIIFDSDWNPHQDLQAQDRAHRIGQQNEVRVLRLCTVNSVEEKIL 834
Query 61 HSAELKLDKDALVIKSGMYN 80
+A+ KL+ D VI++GM++
Sbjct 835 AAAKYKLNVDQKVIQAGMFD 854
> ath:AT2G28290 SYD; SYD (SPLAYED); ATPase/ chromatin binding
Length=3543
Score = 110 bits (274), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 62/138 (44%), Positives = 89/138 (64%), Gaps = 4/138 (2%)
Query 2 KAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQILH 61
+AGG+GVNLQ+ADTVI+FD+DWNPQ D QAQ+RAHRIGQK++VL +RF T +S+EEQ+
Sbjct 1150 RAGGVGVNLQAADTVILFDTDWNPQVDLQAQARAHRIGQKKDVLVLRFETVNSVEEQVRA 1209
Query 62 SAELKLDKDALVIKSGMYNGELQDRETERQEQVREILRRRKQLEANWSRPFDFGILKSQL 121
SAE KL I +G ++ +R+E + +LR K+ E + D L +
Sbjct 1210 SAEHKLGVANQSITAGFFDNNTSAE--DRKEYLESLLRESKKEED--APVLDDDALNDLI 1265
Query 122 ARNPKEATLFDALQKIRQ 139
AR E +F+++ K R+
Sbjct 1266 ARRESEIDIFESIDKQRK 1283
> ath:AT5G19310 homeotic gene regulator, putative; K11647 SWI/SNF-related
matrix-associated actin-dependent regulator of chromatin
subfamily A member 2/4 [EC:3.6.4.-]
Length=1064
Score = 108 bits (271), Expect = 5e-24, Method: Compositional matrix adjust.
Identities = 54/100 (54%), Positives = 73/100 (73%), Gaps = 2/100 (2%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
T+AGGLG+NLQ+ADT+IIFDSDWNPQ D+QA+ RAHRIGQK+EV V+ SIEE IL
Sbjct 771 TRAGGLGLNLQTADTIIIFDSDWNPQMDQQAEDRAHRIGQKKEVRVFVLVSIGSIEEVIL 830
Query 61 HSAELKLDKDALVIKSGMYNGELQDRETERQEQVREILRR 100
A+ K+ DA VI++G++N +R+E + EI+ +
Sbjct 831 ERAKQKMGIDAKVIQAGLFN--TTSTAQDRREMLEEIMSK 868
> mmu:67155 Smarca2, 2610209L14Rik, SNF2alpha, Snf2l2, brahma,
brm; SWI/SNF related, matrix associated, actin dependent regulator
of chromatin, subfamily a, member 2; K11647 SWI/SNF-related
matrix-associated actin-dependent regulator of chromatin
subfamily A member 2/4 [EC:3.6.4.-]
Length=1583
Score = 108 bits (270), Expect = 5e-24, Method: Compositional matrix adjust.
Identities = 49/80 (61%), Positives = 65/80 (81%), Gaps = 0/80 (0%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
T+AGGLG+NLQ+ADTV+IFDSDWNP D QAQ RAHRIGQ+ EV +R T +S+EE+IL
Sbjct 1137 TRAGGLGLNLQAADTVVIFDSDWNPHQDLQAQDRAHRIGQQNEVRVLRLCTVNSVEEKIL 1196
Query 61 HSAELKLDKDALVIKSGMYN 80
+A+ KL+ D VI++GM++
Sbjct 1197 AAAKYKLNVDQKVIQAGMFD 1216
> hsa:6595 SMARCA2, BAF190, BRM, FLJ36757, MGC74511, SNF2, SNF2L2,
SNF2LA, SWI2, Sth1p, hBRM, hSNF2a; SWI/SNF related, matrix
associated, actin dependent regulator of chromatin, subfamily
a, member 2; K11647 SWI/SNF-related matrix-associated
actin-dependent regulator of chromatin subfamily A member 2/4
[EC:3.6.4.-]
Length=1590
Score = 108 bits (270), Expect = 5e-24, Method: Compositional matrix adjust.
Identities = 49/80 (61%), Positives = 65/80 (81%), Gaps = 0/80 (0%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
T+AGGLG+NLQ+ADTV+IFDSDWNP D QAQ RAHRIGQ+ EV +R T +S+EE+IL
Sbjct 1126 TRAGGLGLNLQAADTVVIFDSDWNPHQDLQAQDRAHRIGQQNEVRVLRLCTVNSVEEKIL 1185
Query 61 HSAELKLDKDALVIKSGMYN 80
+A+ KL+ D VI++GM++
Sbjct 1186 AAAKYKLNVDQKVIQAGMFD 1205
> ath:AT2G13370 CHR5; CHR5 (chromatin remodeling 5); ATP binding
/ DNA binding / chromatin binding / helicase/ nucleic acid
binding; K11367 chromodomain-helicase-DNA-binding protein
1 [EC:3.6.4.12]
Length=1724
Score = 108 bits (269), Expect = 8e-24, Method: Compositional matrix adjust.
Identities = 54/90 (60%), Positives = 67/90 (74%), Gaps = 0/90 (0%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
T+AGGLG+NL +ADTV+IFDSDWNPQND QA SRAHRIGQ+ V RFVT S+EE+IL
Sbjct 1015 TRAGGLGINLATADTVVIFDSDWNPQNDLQAMSRAHRIGQQEVVNIYRFVTSKSVEEEIL 1074
Query 61 HSAELKLDKDALVIKSGMYNGELQDRETER 90
A+ K+ D LVI+ G L+ RET++
Sbjct 1075 ERAKRKMVLDHLVIQKLNAEGRLEKRETKK 1104
> xla:100335135 smarca4, baf190, brg1, rtps2, snf2, snf2-beta,
snf2b, snf2l4, snf2lb, swi2; SWI/SNF related, matrix associated,
actin dependent regulator of chromatin, subfamily a, member
4; K11647 SWI/SNF-related matrix-associated actin-dependent
regulator of chromatin subfamily A member 2/4 [EC:3.6.4.-]
Length=1600
Score = 107 bits (268), Expect = 9e-24, Method: Compositional matrix adjust.
Identities = 49/80 (61%), Positives = 64/80 (80%), Gaps = 0/80 (0%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
T+AGGLG+NLQSADTV+IFDSDWNP D QAQ RAHRIG + EV +R T +S+EE+IL
Sbjct 1141 TRAGGLGLNLQSADTVVIFDSDWNPHQDLQAQDRAHRIGPQNEVRVLRLCTVNSVEEKIL 1200
Query 61 HSAELKLDKDALVIKSGMYN 80
+A+ KL+ D VI++GM++
Sbjct 1201 AAAKYKLNVDQKVIQAGMFD 1220
> sce:YOR290C SNF2, GAM1, HAF1, SWI2, TYE3; Catalytic subunit
of the SWI/SNF chromatin remodeling complex involved in transcriptional
regulation; contains DNA-stimulated ATPase activity;
functions interdependently in transcriptional activation
with Snf5p and Snf6p (EC:3.6.1.-); K11786 ATP-dependent helicase
STH1/SNF2 [EC:3.6.4.-]
Length=1703
Score = 107 bits (267), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 52/89 (58%), Positives = 65/89 (73%), Gaps = 0/89 (0%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
T+AGGLG+NLQ+ADTVIIFD+DWNP D QAQ RAHRIGQK EV +R +T +S+EE IL
Sbjct 1163 TRAGGLGLNLQTADTVIIFDTDWNPHQDLQAQDRAHRIGQKNEVRILRLITTNSVEEVIL 1222
Query 61 HSAELKLDKDALVIKSGMYNGELQDRETE 89
A KLD D VI++G ++ + E E
Sbjct 1223 ERAYKKLDIDGKVIQAGKFDNKSTSEEQE 1251
> ath:AT3G06400 CHR11; CHR11 (CHROMATIN-REMODELING PROTEIN 11);
ATP binding / DNA binding / DNA-dependent ATPase/ helicase/
hydrolase, acting on acid anhydrides, in phosphorus-containing
anhydrides / nucleic acid binding / nucleosome binding;
K11654 SWI/SNF-related matrix-associated actin-dependent regulator
of chromatin subfamily A member 5 [EC:3.6.4.-]
Length=1055
Score = 105 bits (261), Expect = 5e-23, Method: Compositional matrix adjust.
Identities = 49/77 (63%), Positives = 60/77 (77%), Gaps = 0/77 (0%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
T+AGGLG+NL +AD VI++DSDWNPQ D QAQ RAHRIGQK+EV RF T S+IEE+++
Sbjct 566 TRAGGLGINLATADVVILYDSDWNPQVDLQAQDRAHRIGQKKEVQVFRFCTESAIEEKVI 625
Query 61 HSAELKLDKDALVIKSG 77
A KL DALVI+ G
Sbjct 626 ERAYKKLALDALVIQQG 642
> cpv:cgd1_3670 CHD3 ortholog with 2x chromodomains plus SNF2
ATpase (2chromo+helicase+Znf_NFX) ; K11367 chromodomain-helicase-DNA-binding
protein 1 [EC:3.6.4.12]
Length=2055
Score = 105 bits (261), Expect = 7e-23, Method: Compositional matrix adjust.
Identities = 51/80 (63%), Positives = 63/80 (78%), Gaps = 1/80 (1%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
TKAGGLG+NL +ADTVII+DSDWNPQND QA++RAHRIGQK++V R VT SIEE IL
Sbjct 948 TKAGGLGINLTTADTVIIYDSDWNPQNDLQAEARAHRIGQKKQVQIYRLVTKDSIEENIL 1007
Query 61 HSAELKLDKDALVIKSGMYN 80
A+ K+ D LV++ G+ N
Sbjct 1008 ERAKTKMVLDTLVVQ-GLNN 1026
> hsa:1105 CHD1, DKFZp686E2337; chromodomain helicase DNA binding
protein 1 (EC:3.6.4.12); K11367 chromodomain-helicase-DNA-binding
protein 1 [EC:3.6.4.12]
Length=1710
Score = 104 bits (260), Expect = 8e-23, Method: Compositional matrix adjust.
Identities = 50/75 (66%), Positives = 60/75 (80%), Gaps = 0/75 (0%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
T+AGGLG+NL SADTV+IFDSDWNPQND QAQ+RAHRIGQK++V R VT S+EE IL
Sbjct 864 TRAGGLGINLASADTVVIFDSDWNPQNDLQAQARAHRIGQKKQVNIYRLVTKGSVEEDIL 923
Query 61 HSAELKLDKDALVIK 75
A+ K+ D LVI+
Sbjct 924 ERAKKKMVLDHLVIQ 938
> dre:563236 MGC173506, wu:fc26h11, wu:fk85d05; zgc:173506; K11367
chromodomain-helicase-DNA-binding protein 1 [EC:3.6.4.12]
Length=1693
Score = 104 bits (260), Expect = 8e-23, Method: Compositional matrix adjust.
Identities = 49/75 (65%), Positives = 61/75 (81%), Gaps = 0/75 (0%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
T+AGGLG+NL SADTV+IFDSDWNPQND QAQ+RAHRIGQK++V R VT S+EE+I+
Sbjct 851 TRAGGLGINLASADTVVIFDSDWNPQNDLQAQARAHRIGQKKQVNIYRLVTKGSVEEEII 910
Query 61 HSAELKLDKDALVIK 75
A+ K+ D LVI+
Sbjct 911 ERAKKKMVLDHLVIQ 925
> mmu:12648 Chd1, 4930525N21Rik, AI851787, AW555109, MGC141554;
chromodomain helicase DNA binding protein 1 (EC:3.6.4.12);
K11367 chromodomain-helicase-DNA-binding protein 1 [EC:3.6.4.12]
Length=1711
Score = 104 bits (260), Expect = 8e-23, Method: Compositional matrix adjust.
Identities = 50/75 (66%), Positives = 60/75 (80%), Gaps = 0/75 (0%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
T+AGGLG+NL SADTV+IFDSDWNPQND QAQ+RAHRIGQK++V R VT S+EE IL
Sbjct 862 TRAGGLGINLASADTVVIFDSDWNPQNDLQAQARAHRIGQKKQVNIYRLVTKGSVEEDIL 921
Query 61 HSAELKLDKDALVIK 75
A+ K+ D LVI+
Sbjct 922 ERAKKKMVLDHLVIQ 936
> dre:282615 smarca5, chunp6878, fb26d12, fb49g04, im:7146484,
wu:fb26d12, wu:fb49g04, zgc:158434; SWI/SNF related, matrix
associated, actin dependent regulator of chromatin, subfamily
a, member 5; K11654 SWI/SNF-related matrix-associated actin-dependent
regulator of chromatin subfamily A member 5 [EC:3.6.4.-]
Length=1028
Score = 103 bits (257), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 46/77 (59%), Positives = 62/77 (80%), Gaps = 0/77 (0%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
T+AGGLG+NL +AD VII+DSDWNPQ D QA RAHRIGQK++V RF+T +++EE+I+
Sbjct 535 TRAGGLGINLATADVVIIYDSDWNPQVDLQAMDRAHRIGQKKQVRVFRFITDNTVEERIV 594
Query 61 HSAELKLDKDALVIKSG 77
AE+KL D++VI+ G
Sbjct 595 ERAEMKLRLDSIVIQQG 611
> mmu:244059 Chd2, 2810013C04Rik, 2810040A01Rik, 5630401D06Rik,
AI851092, BC029703; chromodomain helicase DNA binding protein
2; K11367 chromodomain-helicase-DNA-binding protein 1 [EC:3.6.4.12]
Length=1827
Score = 103 bits (257), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 48/75 (64%), Positives = 61/75 (81%), Gaps = 0/75 (0%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
T+AGGLG+NL SADTV+IFDSDWNPQND QAQ+RAHRIGQK++V R VT ++EE+I+
Sbjct 867 TRAGGLGINLASADTVVIFDSDWNPQNDLQAQARAHRIGQKKQVNIYRLVTKGTVEEEII 926
Query 61 HSAELKLDKDALVIK 75
A+ K+ D LVI+
Sbjct 927 ERAKKKMVLDHLVIQ 941
> tpv:TP01_0866 hypothetical protein; K11367 chromodomain-helicase-DNA-binding
protein 1 [EC:3.6.4.12]
Length=1816
Score = 103 bits (256), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 49/75 (65%), Positives = 59/75 (78%), Gaps = 0/75 (0%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
TKAGGLG+NL SADTVII+DSDWNPQND QA++RAHRIGQ + V R VT SIE+ IL
Sbjct 1192 TKAGGLGINLTSADTVIIYDSDWNPQNDLQAEARAHRIGQTKTVQIYRLVTKDSIEQTIL 1251
Query 61 HSAELKLDKDALVIK 75
A+ K+ DALV++
Sbjct 1252 ERAKTKMVLDALVVQ 1266
> tgo:TGME49_058240 chromodomain helicase DNA binding protein,
putative (EC:2.7.11.1 2.7.7.19); K11367 chromodomain-helicase-DNA-binding
protein 1 [EC:3.6.4.12]
Length=2279
Score = 103 bits (256), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 50/76 (65%), Positives = 58/76 (76%), Gaps = 0/76 (0%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
TKAGGLG+NL SADTVIIFDSDWNPQND QA++RAHRIGQ R V R VT SIE+ IL
Sbjct 1357 TKAGGLGINLTSADTVIIFDSDWNPQNDLQAEARAHRIGQTRTVQIYRLVTKDSIEQTIL 1416
Query 61 HSAELKLDKDALVIKS 76
A+ K+ D LV++
Sbjct 1417 ERAKAKMVLDTLVVQG 1432
> bbo:BBOV_IV009080 23.m05963; chromo-helicase DNA-binding protein;
K11367 chromodomain-helicase-DNA-binding protein 1 [EC:3.6.4.12]
Length=1729
Score = 102 bits (253), Expect = 6e-22, Method: Compositional matrix adjust.
Identities = 48/76 (63%), Positives = 59/76 (77%), Gaps = 0/76 (0%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
TKAGGLG+NL +ADTVII+DSDWNPQND QA++RAHRIGQ + V R VT SIE+ IL
Sbjct 1116 TKAGGLGINLTTADTVIIYDSDWNPQNDLQAEARAHRIGQTKTVQIYRLVTKDSIEQTIL 1175
Query 61 HSAELKLDKDALVIKS 76
A+ K+ DALV++
Sbjct 1176 ERAKTKMVLDALVVQG 1191
> cel:F26F12.7 let-418; LEThal family member (let-418); K11643
chromodomain-helicase-DNA-binding protein 4 [EC:3.6.4.12]
Length=1829
Score = 101 bits (252), Expect = 6e-22, Method: Compositional matrix adjust.
Identities = 46/78 (58%), Positives = 62/78 (79%), Gaps = 0/78 (0%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
T+AGGLG+NL +ADTVII+DSDWNP ND QA SRAHR+GQK +V+ RFVT S+EE+I
Sbjct 1002 TRAGGLGINLATADTVIIYDSDWNPHNDIQAFSRAHRLGQKHKVMIYRFVTKKSVEEKIT 1061
Query 61 HSAELKLDKDALVIKSGM 78
A+ K+ + LV+++G+
Sbjct 1062 SVAKKKMLLNHLVVRAGL 1079
> sce:YOR304W ISW2; ATP-dependent DNA translocase involved in
chromatin remodeling; ATPase component that, with Itc1p, forms
a complex required for repression of A-specific genes, INO1,
and early meiotic genes during mitotic growth (EC:3.6.1.-)
Length=1120
Score = 101 bits (252), Expect = 6e-22, Method: Compositional matrix adjust.
Identities = 48/77 (62%), Positives = 60/77 (77%), Gaps = 0/77 (0%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
T+AGGLG+NL +ADTVI+FDSDWNPQ D QA RAHRIGQK++V RFVT ++IEE+++
Sbjct 566 TRAGGLGINLVTADTVILFDSDWNPQADLQAMDRAHRIGQKKQVHVYRFVTENAIEEKVI 625
Query 61 HSAELKLDKDALVIKSG 77
A KL D LVI+ G
Sbjct 626 ERAAQKLRLDQLVIQQG 642
> ath:AT5G18620 CHR17; CHR17 (CHROMATIN REMODELING FACTOR17);
ATP binding / DNA binding / DNA-dependent ATPase/ helicase/
hydrolase, acting on acid anhydrides, in phosphorus-containing
anhydrides / nucleic acid binding / nucleosome binding; K11654
SWI/SNF-related matrix-associated actin-dependent regulator
of chromatin subfamily A member 5 [EC:3.6.4.-]
Length=1069
Score = 101 bits (251), Expect = 8e-22, Method: Compositional matrix adjust.
Identities = 47/77 (61%), Positives = 59/77 (76%), Gaps = 0/77 (0%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
T+AGGLG+NL +AD VI++DSDWNPQ D QAQ RAHRIGQK+EV RF T ++IE +++
Sbjct 571 TRAGGLGINLATADVVILYDSDWNPQVDLQAQDRAHRIGQKKEVQVFRFCTENAIEAKVI 630
Query 61 HSAELKLDKDALVIKSG 77
A KL DALVI+ G
Sbjct 631 ERAYKKLALDALVIQQG 647
> cel:H06O01.2 hypothetical protein; K11367 chromodomain-helicase-DNA-binding
protein 1 [EC:3.6.4.12]
Length=1461
Score = 100 bits (250), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 49/75 (65%), Positives = 58/75 (77%), Gaps = 0/75 (0%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
T+AGGLG+NL +ADTVIIFDSDWNPQND QA SRAHRIGQ + V R VT S+EE+I+
Sbjct 787 TRAGGLGINLATADTVIIFDSDWNPQNDLQAMSRAHRIGQTKTVNIYRLVTKGSVEEEIV 846
Query 61 HSAELKLDKDALVIK 75
A+ KL D LVI+
Sbjct 847 ERAKRKLVLDHLVIQ 861
> xla:380196 chd4, MGC52739, b230399n07; chromodomain helicase
DNA binding protein 4; K11643 chromodomain-helicase-DNA-binding
protein 4 [EC:3.6.4.12]
Length=1893
Score = 100 bits (250), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 46/78 (58%), Positives = 61/78 (78%), Gaps = 0/78 (0%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
T+AGGLG+NL +ADTV+I+DSDWNP ND QA SRAHRIGQ R+V+ RFVT +S+EE+I
Sbjct 1117 TRAGGLGINLATADTVVIYDSDWNPHNDIQAFSRAHRIGQNRKVMIYRFVTRASVEERIT 1176
Query 61 HSAELKLDKDALVIKSGM 78
A+ K+ LV++ G+
Sbjct 1177 QVAKKKMMLTHLVVRPGL 1194
> cel:T14G8.1 chd-3; CHromoDomain protein family member (chd-3)
Length=1787
Score = 100 bits (250), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 46/78 (58%), Positives = 61/78 (78%), Gaps = 0/78 (0%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
T+AGGLG+NL +ADTVII+DSDWNP ND QA SRAHR+GQK +V+ RFVT S+EE+I
Sbjct 1016 TRAGGLGINLATADTVIIYDSDWNPHNDIQAFSRAHRLGQKHKVMIYRFVTKGSVEERIT 1075
Query 61 HSAELKLDKDALVIKSGM 78
A+ K+ LV+++G+
Sbjct 1076 SVAKKKMLLTHLVVRAGL 1093
> xla:399165 smarca5, iswi; SWI/SNF related, matrix associated,
actin dependent regulator of chromatin, subfamily a, member
5; K11654 SWI/SNF-related matrix-associated actin-dependent
regulator of chromatin subfamily A member 5 [EC:3.6.4.-]
Length=1046
Score = 100 bits (250), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 45/77 (58%), Positives = 60/77 (77%), Gaps = 0/77 (0%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
T+AGGLG+NL +AD VII+DSDWNPQ D QA RAHRIGQ + V RF+T +++EE+I+
Sbjct 552 TRAGGLGINLATADVVIIYDSDWNPQVDLQAMDRAHRIGQTKTVRVFRFITDNTVEERIV 611
Query 61 HSAELKLDKDALVIKSG 77
AE+KL D++VI+ G
Sbjct 612 ERAEMKLRLDSIVIQQG 628
> dre:560622 fd12d03, wu:fb44b12; wu:fd12d03
Length=1953
Score = 100 bits (250), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 46/78 (58%), Positives = 61/78 (78%), Gaps = 0/78 (0%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
T+AGGLG+NL +ADTVII+DSDWNP ND QA SRAHRIGQ ++V+ RFVT +S+EE+I
Sbjct 1136 TRAGGLGINLATADTVIIYDSDWNPHNDIQAFSRAHRIGQNKKVMIYRFVTKASVEERIT 1195
Query 61 HSAELKLDKDALVIKSGM 78
A+ K+ LV++ G+
Sbjct 1196 QVAKKKMMLTHLVVRPGL 1213
> pfa:PF11_0053 PfSNF2L; K01509 adenosinetriphosphatase [EC:3.6.1.3]
Length=1426
Score = 100 bits (250), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 49/98 (50%), Positives = 70/98 (71%), Gaps = 2/98 (2%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
T+AGG+G+NL +AD VI+FDSD+NPQ D QA RAHRIGQK+ V+ RFVT +S+EE+I+
Sbjct 702 TRAGGIGINLTTADIVILFDSDYNPQMDIQAMDRAHRIGQKKRVIVYRFVTQNSVEEKIV 761
Query 61 HSAELKLDKDALVIKSGMYNGELQDRETERQEQVREIL 98
A KL D+L+I+ G N L + ++++ +IL
Sbjct 762 ERAAKKLKLDSLIIQKGKLN--LNSAKENNKQELHDIL 797
> mmu:93761 Smarca1, 5730494M04Rik, Snf2l; SWI/SNF related, matrix
associated, actin dependent regulator of chromatin, subfamily
a, member 1; K11727 SWI/SNF-related matrix-associated
actin-dependent regulator of chromatin subfamily A member 1
[EC:3.6.4.-]
Length=1046
Score = 100 bits (249), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 45/77 (58%), Positives = 60/77 (77%), Gaps = 0/77 (0%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
T+AGGLG+NL SAD VI++DSDWNPQ D QA RAHRIGQK+ V R +T +++EE+I+
Sbjct 566 TRAGGLGINLASADVVILYDSDWNPQVDLQAMDRAHRIGQKKPVRVFRLITDNTVEERIV 625
Query 61 HSAELKLDKDALVIKSG 77
AE+KL D++VI+ G
Sbjct 626 ERAEIKLRLDSIVIQQG 642
> hsa:6594 SMARCA1, DKFZp686D1623, FLJ41547, ISWI, NURF140, SNF2L,
SNF2L1, SNF2LB, SNF2LT, SWI, SWI2; SWI/SNF related, matrix
associated, actin dependent regulator of chromatin, subfamily
a, member 1; K11727 SWI/SNF-related matrix-associated
actin-dependent regulator of chromatin subfamily A member 1
[EC:3.6.4.-]
Length=1054
Score = 100 bits (249), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 45/77 (58%), Positives = 60/77 (77%), Gaps = 0/77 (0%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
T+AGGLG+NL SAD VI++DSDWNPQ D QA RAHRIGQK+ V R +T +++EE+I+
Sbjct 574 TRAGGLGINLASADVVILYDSDWNPQVDLQAMDRAHRIGQKKPVRVFRLITDNTVEERIV 633
Query 61 HSAELKLDKDALVIKSG 77
AE+KL D++VI+ G
Sbjct 634 ERAEIKLRLDSIVIQQG 650
> mmu:216848 Chd3, 2600010P09Rik, AF020312, Chd7, MGC40857, Prp7,
Prp9-1; chromodomain helicase DNA binding protein 3; K11642
chromodomain-helicase-DNA-binding protein 3 [EC:3.6.4.12]
Length=2021
Score = 100 bits (249), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 47/78 (60%), Positives = 60/78 (76%), Gaps = 0/78 (0%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
T+AGGLG+NL +ADTVIIFDSDWNP ND QA SRAHRIGQ +V+ RFVT +S+EE+I
Sbjct 1188 TRAGGLGINLATADTVIIFDSDWNPHNDIQAFSRAHRIGQANKVMIYRFVTRASVEERIT 1247
Query 61 HSAELKLDKDALVIKSGM 78
A+ K+ LV++ G+
Sbjct 1248 QVAKRKMMLTHLVVRPGL 1265
> dre:568230 similar to CHD3; K11642 chromodomain-helicase-DNA-binding
protein 3 [EC:3.6.4.12]
Length=2063
Score = 100 bits (249), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 47/78 (60%), Positives = 60/78 (76%), Gaps = 0/78 (0%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
T+AGGLG+NL +ADTVIIFDSDWNP ND QA SRAHRIGQ +V+ RFVT +S+EE+I
Sbjct 1151 TRAGGLGINLATADTVIIFDSDWNPHNDIQAFSRAHRIGQANKVMIYRFVTRASVEERIT 1210
Query 61 HSAELKLDKDALVIKSGM 78
A+ K+ LV++ G+
Sbjct 1211 QVAKRKMMLTHLVVRPGL 1228
> mmu:269610 Chd5, 4930532L22Rik, AW060752, B230399N07Rik; chromodomain
helicase DNA binding protein 5; K14435 chromodomain-helicase-DNA-binding
protein 5 [EC:3.6.4.12]
Length=1952
Score = 100 bits (249), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 46/78 (58%), Positives = 61/78 (78%), Gaps = 0/78 (0%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
T+AGGLG+NL +ADTVII+DSDWNP ND QA SRAHRIGQ ++V+ RFVT +S+EE+I
Sbjct 1102 TRAGGLGINLATADTVIIYDSDWNPHNDIQAFSRAHRIGQNKKVMIYRFVTRASVEERIT 1161
Query 61 HSAELKLDKDALVIKSGM 78
A+ K+ LV++ G+
Sbjct 1162 QVAKRKMMLTHLVVRPGL 1179
> hsa:1107 CHD3, Mi-2a, Mi2-ALPHA, ZFH; chromodomain helicase
DNA binding protein 3 (EC:3.6.4.12); K11642 chromodomain-helicase-DNA-binding
protein 3 [EC:3.6.4.12]
Length=2059
Score = 100 bits (249), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 47/78 (60%), Positives = 60/78 (76%), Gaps = 0/78 (0%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
T+AGGLG+NL +ADTVIIFDSDWNP ND QA SRAHRIGQ +V+ RFVT +S+EE+I
Sbjct 1195 TRAGGLGINLATADTVIIFDSDWNPHNDIQAFSRAHRIGQANKVMIYRFVTRASVEERIT 1254
Query 61 HSAELKLDKDALVIKSGM 78
A+ K+ LV++ G+
Sbjct 1255 QVAKRKMMLTHLVVRPGL 1272
> hsa:26038 CHD5, DKFZp434N231, KIAA0444; chromodomain helicase
DNA binding protein 5 (EC:3.6.4.12); K14435 chromodomain-helicase-DNA-binding
protein 5 [EC:3.6.4.12]
Length=1954
Score = 100 bits (249), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 46/78 (58%), Positives = 61/78 (78%), Gaps = 0/78 (0%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
T+AGGLG+NL +ADTVII+DSDWNP ND QA SRAHRIGQ ++V+ RFVT +S+EE+I
Sbjct 1100 TRAGGLGINLATADTVIIYDSDWNPHNDIQAFSRAHRIGQNKKVMIYRFVTRASVEERIT 1159
Query 61 HSAELKLDKDALVIKSGM 78
A+ K+ LV++ G+
Sbjct 1160 QVAKRKMMLTHLVVRPGL 1177
> hsa:1108 CHD4, DKFZp686E06161, Mi-2b, Mi2-BETA; chromodomain
helicase DNA binding protein 4 (EC:3.6.4.12); K11643 chromodomain-helicase-DNA-binding
protein 4 [EC:3.6.4.12]
Length=1912
Score = 100 bits (249), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 46/78 (58%), Positives = 61/78 (78%), Gaps = 0/78 (0%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
T+AGGLG+NL +ADTVII+DSDWNP ND QA SRAHRIGQ ++V+ RFVT +S+EE+I
Sbjct 1126 TRAGGLGINLATADTVIIYDSDWNPHNDIQAFSRAHRIGQNKKVMIYRFVTRASVEERIT 1185
Query 61 HSAELKLDKDALVIKSGM 78
A+ K+ LV++ G+
Sbjct 1186 QVAKKKMMLTHLVVRPGL 1203
> dre:558344 im:7143343; si:ch211-51m24.3; K11643 chromodomain-helicase-DNA-binding
protein 4 [EC:3.6.4.12]
Length=1929
Score = 100 bits (249), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 45/78 (57%), Positives = 61/78 (78%), Gaps = 0/78 (0%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
T+AGGLG+NL +ADTV+I+DSDWNP ND QA SRAHRIGQ ++V+ RFVT +S+EE+I
Sbjct 1111 TRAGGLGINLATADTVVIYDSDWNPHNDIQAFSRAHRIGQNKKVMIYRFVTKASVEERIT 1170
Query 61 HSAELKLDKDALVIKSGM 78
A+ K+ LV++ G+
Sbjct 1171 QVAKKKMMLTHLVVRPGL 1188
> cel:T04D1.4 tag-192; Temporarily Assigned Gene name family member
(tag-192); K14437 chromodomain-helicase-DNA-binding protein
7 [EC:3.6.4.12]
Length=2967
Score = 100 bits (248), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 57/129 (44%), Positives = 73/129 (56%), Gaps = 27/129 (20%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
T+AGGLG+NL +ADTVIIFDSDWNPQND QAQ+R HRIGQK+ V R +T ++ E ++
Sbjct 1597 TRAGGLGINLTAADTVIIFDSDWNPQNDLQAQARCHRIGQKKLVKVYRLITSNTYEREMF 1656
Query 61 HSAELKLDKDALVIKS----------------------GMYNGELQDRETE----RQEQV 94
A LKL D V++S G Y G + D E E +E +
Sbjct 1657 DKASLKLGLDKAVLQSTTALKAEGTALSKKDVEELLKKGAY-GSIMDEENESSKFNEEDI 1715
Query 95 REILRRRKQ 103
IL+RR Q
Sbjct 1716 ETILQRRTQ 1724
> mmu:107932 Chd4, 9530019N15Rik, AA617397, BC005710, D6Ertd380e,
KIAA4075, MGC11769, Mi-2beta, mKIAA4075; chromodomain helicase
DNA binding protein 4 (EC:3.6.4.12); K11643 chromodomain-helicase-DNA-binding
protein 4 [EC:3.6.4.12]
Length=1915
Score = 100 bits (248), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 46/78 (58%), Positives = 61/78 (78%), Gaps = 0/78 (0%)
Query 1 TKAGGLGVNLQSADTVIIFDSDWNPQNDEQAQSRAHRIGQKREVLTIRFVTPSSIEEQIL 60
T+AGGLG+NL +ADTVII+DSDWNP ND QA SRAHRIGQ ++V+ RFVT +S+EE+I
Sbjct 1119 TRAGGLGINLATADTVIIYDSDWNPHNDIQAFSRAHRIGQNKKVMIYRFVTRASVEERIT 1178
Query 61 HSAELKLDKDALVIKSGM 78
A+ K+ LV++ G+
Sbjct 1179 QVAKKKMMLTHLVVRPGL 1196
Lambda K H
0.317 0.133 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2749206264
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40