bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2579_orf1
Length=145
Score E
Sequences producing significant alignments: (Bits) Value
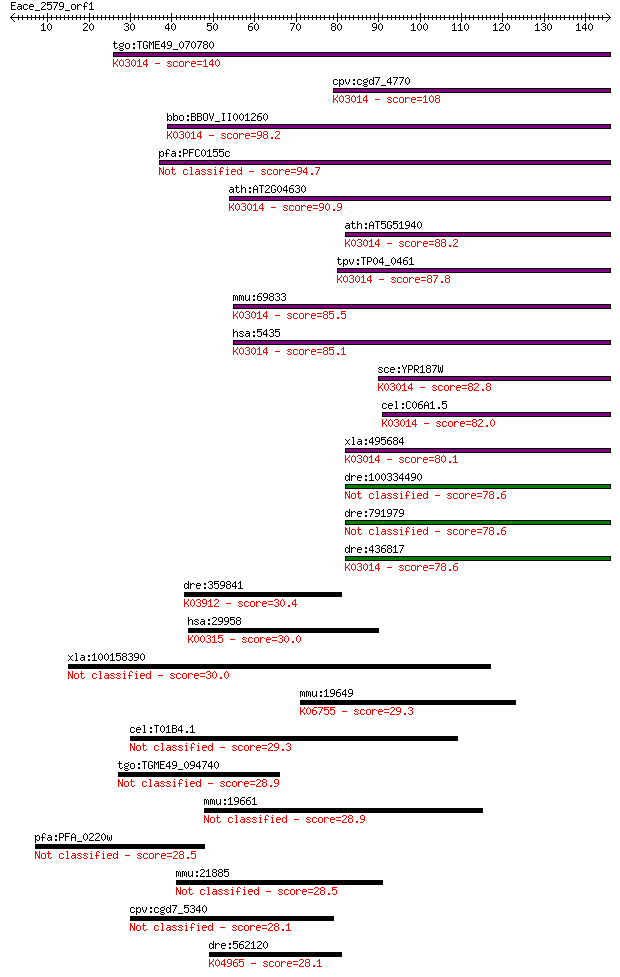
tgo:TGME49_070780 RNA polymerase II subunit RPB6, putative (EC... 140 9e-34
cpv:cgd7_4770 DNA-directed RNA polymerase subunit ; K03014 DNA... 108 8e-24
bbo:BBOV_II001260 18.m06094; DNA-directed RNA polymerase subun... 98.2 9e-21
pfa:PFC0155c DNA-directed RNA polymerase subunit I, putative (... 94.7 8e-20
ath:AT2G04630 NRPB6B; NRPB6B; DNA binding / DNA-directed RNA p... 90.9 1e-18
ath:AT5G51940 NRPB6A; NRPB6A; DNA binding / DNA-directed RNA p... 88.2 7e-18
tpv:TP04_0461 hypothetical protein; K03014 DNA-directed RNA po... 87.8 9e-18
mmu:69833 Polr2f, 1810060D16Rik; polymerase (RNA) II (DNA dire... 85.5 6e-17
hsa:5435 POLR2F, HRBP14.4, POLRF, RPABC2, RPB14.4, RPB6; polym... 85.1 6e-17
sce:YPR187W RPO26, RPB6; ABC23; K03014 DNA-directed RNA polyme... 82.8 4e-16
cel:C06A1.5 rpb-6; RNA Polymerase II (B) subunit family member... 82.0 6e-16
xla:495684 polr2f; polymerase (RNA) II (DNA directed) polypept... 80.1 2e-15
dre:100334490 DNA directed RNA polymerase II polypeptide F-like 78.6 6e-15
dre:791979 DNA directed RNA polymerase II polypeptide F-like 78.6 7e-15
dre:436817 zgc:92790; K03014 DNA-directed RNA polymerases I, I... 78.6 7e-15
dre:359841 serpind1, HCII; serine (or cysteine) proteinase inh... 30.4 1.8
hsa:29958 DMGDH, DMGDHD, ME2GLYDH; dimethylglycine dehydrogena... 30.0 2.6
xla:100158390 KIAA1841 30.0 2.7
mmu:19649 Robo3, Rbig1, Rig-1, Rig1, Robo3a, Robo3b; roundabou... 29.3 4.3
cel:T01B4.1 twk-21; TWiK family of potassium channels family m... 29.3 4.8
tgo:TGME49_094740 armadillo/beta-catenin-like repeat-containin... 28.9 5.2
mmu:19661 Rbp3, Irbp, MGC102323, Rbp-3; retinol binding protei... 28.9 5.9
pfa:PFA_0220w ubiquitin carboxyl-terminal hydrolase, putative ... 28.5 6.8
mmu:21885 Tle1, C230057C06Rik, Estm14, Grg1, Tle4l; transducin... 28.5 7.9
cpv:cgd7_5340 hypothetical protein 28.1 9.6
dre:562120 trpc2; transient receptor potential cation channel,... 28.1 9.6
> tgo:TGME49_070780 RNA polymerase II subunit RPB6, putative (EC:2.7.7.6);
K03014 DNA-directed RNA polymerases I, II, and
III subunit RPABC2
Length=146
Score = 140 bits (354), Expect = 9e-34, Method: Compositional matrix adjust.
Identities = 78/126 (61%), Positives = 91/126 (72%), Gaps = 6/126 (4%)
Query 26 MADDENDHMFAEPAFGDEFGDELGEEDL----DEDIEEADAAA--EDVDVVIDPNDPRLQ 79
MADDE DHMFA G D D D+E A AAA E ++DP+DPR
Sbjct 1 MADDEIDHMFAGEPGGLGDDFGEDFGDDELIDDFDVERAAAAASHEPESDILDPSDPRAA 60
Query 80 RPEAHPSEGPRITTPFLTKFEKARIIGTRALQISMNAPLTIQLEGESDPLIIAEKELYQK 139
RP A PSEGPRIT+P++TKFEKAR+IGTRALQISMNAP+TI L+GE+DPLIIAEKELYQK
Sbjct 61 RPHARPSEGPRITSPYITKFEKARVIGTRALQISMNAPITIPLDGETDPLIIAEKELYQK 120
Query 140 TLPFIV 145
T+PF +
Sbjct 121 TIPFTI 126
> cpv:cgd7_4770 DNA-directed RNA polymerase subunit ; K03014 DNA-directed
RNA polymerases I, II, and III subunit RPABC2
Length=129
Score = 108 bits (269), Expect = 8e-24, Method: Compositional matrix adjust.
Identities = 48/67 (71%), Positives = 59/67 (88%), Gaps = 0/67 (0%)
Query 79 QRPEAHPSEGPRITTPFLTKFEKARIIGTRALQISMNAPLTIQLEGESDPLIIAEKELYQ 138
R + P+EGPRITTP++TKFEKARIIGTRALQISMNAP+ I L+GE+DPL+IAEKELY
Sbjct 43 SRYDGKPNEGPRITTPYMTKFEKARIIGTRALQISMNAPVAIPLDGETDPLLIAEKELYT 102
Query 139 KTLPFIV 145
K +PF++
Sbjct 103 KKIPFVI 109
> bbo:BBOV_II001260 18.m06094; DNA-directed RNA polymerase subunit
I; K03014 DNA-directed RNA polymerases I, II, and III subunit
RPABC2
Length=154
Score = 98.2 bits (243), Expect = 9e-21, Method: Compositional matrix adjust.
Identities = 62/136 (45%), Positives = 83/136 (61%), Gaps = 34/136 (25%)
Query 39 AFGDEFGDELG-------EEDLDEDIEEADAAAEDVDVVIDPNDPRLQRPEAHPSEGPRI 91
A+GD+FG+ G EE+ DE + DVD++ DPN+ R + AH RI
Sbjct 4 AYGDDFGENFGIDEFADEEEEYDEFDDYDQQRVADVDIITDPNEYRQRVENAH-----RI 58
Query 92 TTPFLTKFEKARIIGTRALQISMNAPLTIQLE--------------GES--------DPL 129
T+P++TK+EKARIIGTRALQIS+NAP+TI ++ GES DPL
Sbjct 59 TSPYMTKYEKARIIGTRALQISLNAPITIPVDASMDTVDEGITMGFGESVDTSAAAIDPL 118
Query 130 IIAEKELYQKTLPFIV 145
+IAEKELYQK++PFI+
Sbjct 119 VIAEKELYQKSVPFII 134
> pfa:PFC0155c DNA-directed RNA polymerase subunit I, putative
(EC:2.7.7.6)
Length=153
Score = 94.7 bits (234), Expect = 8e-20, Method: Compositional matrix adjust.
Identities = 55/128 (42%), Positives = 74/128 (57%), Gaps = 26/128 (20%)
Query 37 EPAFGDEFGDELGEEDLDEDIEEADAAAEDVDVVIDPNDPRLQRPEAHPSEGP----RIT 92
E GDEFG + D ++ + D+D++ D + + + SE RIT
Sbjct 13 EDFMGDEFGQGI-------DSDDGENYENDIDIITDHQIKKDNKSDYENSEANEDNIRIT 65
Query 93 TPFLTKFEKARIIGTRALQISMNAPLTIQLEGE---------------SDPLIIAEKELY 137
+P+LTK+EKARIIGTRALQISMNAPLTI +E +DPL+IAEKELY
Sbjct 66 SPYLTKYEKARIIGTRALQISMNAPLTIPIETSNDMINSKNEYDNYLNNDPLVIAEKELY 125
Query 138 QKTLPFIV 145
K++PFI+
Sbjct 126 NKSIPFIL 133
> ath:AT2G04630 NRPB6B; NRPB6B; DNA binding / DNA-directed RNA
polymerase; K03014 DNA-directed RNA polymerases I, II, and
III subunit RPABC2
Length=144
Score = 90.9 bits (224), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 48/92 (52%), Positives = 65/92 (70%), Gaps = 7/92 (7%)
Query 54 DEDIEEADAAAEDVDVVIDPNDPRLQRPEAHPSEGPRITTPFLTKFEKARIIGTRALQIS 113
D DI+E D DV +DP + + E P + PR T+ F+TK+E+ARI+GTRALQIS
Sbjct 30 DADIKEND------DVNVDPLETE-DKVETEPVQRPRKTSKFMTKYERARILGTRALQIS 82
Query 114 MNAPLTIQLEGESDPLIIAEKELYQKTLPFIV 145
MNAP+ ++LEGE+DPL IA KEL Q+ +PF +
Sbjct 83 MNAPVMVELEGETDPLEIAMKELRQRKIPFTI 114
> ath:AT5G51940 NRPB6A; NRPB6A; DNA binding / DNA-directed RNA
polymerase; K03014 DNA-directed RNA polymerases I, II, and
III subunit RPABC2
Length=144
Score = 88.2 bits (217), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 39/64 (60%), Positives = 52/64 (81%), Gaps = 0/64 (0%)
Query 82 EAHPSEGPRITTPFLTKFEKARIIGTRALQISMNAPLTIQLEGESDPLIIAEKELYQKTL 141
E P + PR T+ F+TK+E+ARI+GTRALQISMNAP+ ++LEGE+DPL IA KEL Q+ +
Sbjct 51 ETEPVQRPRKTSKFMTKYERARILGTRALQISMNAPVMVELEGETDPLEIAMKELRQRKI 110
Query 142 PFIV 145
PF +
Sbjct 111 PFTI 114
> tpv:TP04_0461 hypothetical protein; K03014 DNA-directed RNA
polymerases I, II, and III subunit RPABC2
Length=391
Score = 87.8 bits (216), Expect = 9e-18, Method: Compositional matrix adjust.
Identities = 49/87 (56%), Positives = 57/87 (65%), Gaps = 26/87 (29%)
Query 80 RPEAHPSEGPRITTPFLTKFEKARIIGTRALQISMNAPLTIQLEGES------------- 126
R AH RIT+P+LTK+EKARIIGTRALQIS+NAP+TI ++G S
Sbjct 290 RLSAH-----RITSPYLTKYEKARIIGTRALQISLNAPITIPIDGPSEATDDGFTTLGDF 344
Query 127 --------DPLIIAEKELYQKTLPFIV 145
DPLIIAEKELYQKT+PFIV
Sbjct 345 GDTQTTAIDPLIIAEKELYQKTVPFIV 371
> mmu:69833 Polr2f, 1810060D16Rik; polymerase (RNA) II (DNA directed)
polypeptide F (EC:2.7.7.6); K03014 DNA-directed RNA
polymerases I, II, and III subunit RPABC2
Length=127
Score = 85.5 bits (210), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 43/92 (46%), Positives = 68/92 (73%), Gaps = 9/92 (9%)
Query 55 EDIEEADA-AAEDVDVVIDPNDPRLQRPEAHPSEGPRITTPFLTKFEKARIIGTRALQIS 113
+D+E A+ E+V+++ P +RP+A+ RITTP++TK+E+AR++GTRALQI+
Sbjct 23 DDLENAEEEGQENVEIL-----PSGERPQANQK---RITTPYMTKYERARVLGTRALQIA 74
Query 114 MNAPLTIQLEGESDPLIIAEKELYQKTLPFIV 145
M AP+ ++LEGE+DPL+IA KEL + +P I+
Sbjct 75 MCAPVMVELEGETDPLLIAMKELKARKIPIII 106
> hsa:5435 POLR2F, HRBP14.4, POLRF, RPABC2, RPB14.4, RPB6; polymerase
(RNA) II (DNA directed) polypeptide F (EC:2.7.7.6);
K03014 DNA-directed RNA polymerases I, II, and III subunit RPABC2
Length=127
Score = 85.1 bits (209), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 43/92 (46%), Positives = 68/92 (73%), Gaps = 9/92 (9%)
Query 55 EDIEEADA-AAEDVDVVIDPNDPRLQRPEAHPSEGPRITTPFLTKFEKARIIGTRALQIS 113
+D+E A+ E+V+++ P +RP+A+ RITTP++TK+E+AR++GTRALQI+
Sbjct 23 DDLENAEEEGQENVEIL-----PSGERPQANQK---RITTPYMTKYERARVLGTRALQIA 74
Query 114 MNAPLTIQLEGESDPLIIAEKELYQKTLPFIV 145
M AP+ ++LEGE+DPL+IA KEL + +P I+
Sbjct 75 MCAPVMVELEGETDPLLIAMKELKARKIPIII 106
> sce:YPR187W RPO26, RPB6; ABC23; K03014 DNA-directed RNA polymerases
I, II, and III subunit RPABC2
Length=155
Score = 82.8 bits (203), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 36/56 (64%), Positives = 48/56 (85%), Gaps = 0/56 (0%)
Query 90 RITTPFLTKFEKARIIGTRALQISMNAPLTIQLEGESDPLIIAEKELYQKTLPFIV 145
R TTP++TK+E+ARI+GTRALQISMNAP+ + LEGE+DPL IA KEL +K +P ++
Sbjct 79 RATTPYMTKYERARILGTRALQISMNAPVFVDLEGETDPLRIAMKELAEKKIPLVI 134
> cel:C06A1.5 rpb-6; RNA Polymerase II (B) subunit family member
(rpb-6); K03014 DNA-directed RNA polymerases I, II, and III
subunit RPABC2
Length=137
Score = 82.0 bits (201), Expect = 6e-16, Method: Compositional matrix adjust.
Identities = 35/55 (63%), Positives = 48/55 (87%), Gaps = 0/55 (0%)
Query 91 ITTPFLTKFEKARIIGTRALQISMNAPLTIQLEGESDPLIIAEKELYQKTLPFIV 145
+TTPF+TK+E+AR++GTRALQI+M AP+ ++LEGE+DPL IA KEL Q+ +P IV
Sbjct 61 VTTPFMTKYERARVLGTRALQIAMGAPVMVELEGETDPLEIARKELKQRRIPIIV 115
> xla:495684 polr2f; polymerase (RNA) II (DNA directed) polypeptide
F (EC:2.7.7.6); K03014 DNA-directed RNA polymerases I,
II, and III subunit RPABC2
Length=127
Score = 80.1 bits (196), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 34/64 (53%), Positives = 50/64 (78%), Gaps = 0/64 (0%)
Query 82 EAHPSEGPRITTPFLTKFEKARIIGTRALQISMNAPLTIQLEGESDPLIIAEKELYQKTL 141
E + RITT ++TK+E+AR++GTRALQI+M AP+ ++LEGE+DPL+IA KEL + +
Sbjct 43 EGQQANQKRITTNYMTKYERARVLGTRALQIAMCAPVMVELEGETDPLLIAMKELKARKI 102
Query 142 PFIV 145
P I+
Sbjct 103 PIII 106
> dre:100334490 DNA directed RNA polymerase II polypeptide F-like
Length=230
Score = 78.6 bits (192), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 34/64 (53%), Positives = 49/64 (76%), Gaps = 0/64 (0%)
Query 82 EAHPSEGPRITTPFLTKFEKARIIGTRALQISMNAPLTIQLEGESDPLIIAEKELYQKTL 141
E + RITT ++TK+E+AR++GTRALQI+M AP+ ++LEGE+DPL IA KEL + +
Sbjct 146 EGQQANQKRITTQYMTKYERARVLGTRALQIAMCAPVMVELEGETDPLQIAMKELKSRKI 205
Query 142 PFIV 145
P I+
Sbjct 206 PIII 209
> dre:791979 DNA directed RNA polymerase II polypeptide F-like
Length=127
Score = 78.6 bits (192), Expect = 7e-15, Method: Compositional matrix adjust.
Identities = 34/64 (53%), Positives = 49/64 (76%), Gaps = 0/64 (0%)
Query 82 EAHPSEGPRITTPFLTKFEKARIIGTRALQISMNAPLTIQLEGESDPLIIAEKELYQKTL 141
E + RITT ++TK+E+AR++GTRALQI+M AP+ ++LEGE+DPL IA KEL + +
Sbjct 43 EGQQANQKRITTQYMTKYERARVLGTRALQIAMCAPVMVELEGETDPLQIAMKELKSRKI 102
Query 142 PFIV 145
P I+
Sbjct 103 PIII 106
> dre:436817 zgc:92790; K03014 DNA-directed RNA polymerases I,
II, and III subunit RPABC2
Length=127
Score = 78.6 bits (192), Expect = 7e-15, Method: Compositional matrix adjust.
Identities = 34/64 (53%), Positives = 49/64 (76%), Gaps = 0/64 (0%)
Query 82 EAHPSEGPRITTPFLTKFEKARIIGTRALQISMNAPLTIQLEGESDPLIIAEKELYQKTL 141
E + RITT ++TK+E+AR++GTRALQI+M AP+ ++LEGE+DPL IA KEL + +
Sbjct 43 EGQQANQKRITTQYMTKYERARVLGTRALQIAMCAPVMVELEGETDPLQIAMKELKSRKI 102
Query 142 PFIV 145
P I+
Sbjct 103 PIII 106
> dre:359841 serpind1, HCII; serine (or cysteine) proteinase inhibitor,
clade D (heparin cofactor), member 1; K03912 heparin
cofactor II
Length=507
Score = 30.4 bits (67), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 15/40 (37%), Positives = 25/40 (62%), Gaps = 2/40 (5%)
Query 43 EFGDELGEEDLDE--DIEEADAAAEDVDVVIDPNDPRLQR 80
+F LGE+D E I+E A D+D+ +P+DP+++R
Sbjct 79 DFDKILGEDDYSEGDHIDEISTPAPDLDLFYEPSDPKIRR 118
> hsa:29958 DMGDH, DMGDHD, ME2GLYDH; dimethylglycine dehydrogenase
(EC:1.5.99.2); K00315 dimethylglycine dehydrogenase [EC:1.5.99.2]
Length=866
Score = 30.0 bits (66), Expect = 2.6, Method: Composition-based stats.
Identities = 20/52 (38%), Positives = 24/52 (46%), Gaps = 6/52 (11%)
Query 44 FGDELGEEDLDEDIEEADAAAEDV------DVVIDPNDPRLQRPEAHPSEGP 89
FG EL E DLD +E AA E V D++ N P P+ P GP
Sbjct 333 FGKELFESDLDRIMEHIKAAMEMVPVLKKADIINVVNGPITYSPDILPMVGP 384
> xla:100158390 KIAA1841
Length=715
Score = 30.0 bits (66), Expect = 2.7, Method: Composition-based stats.
Identities = 20/104 (19%), Positives = 44/104 (42%), Gaps = 5/104 (4%)
Query 15 KNTGLGFRVLKMADDENDHMFAEPAFGDEFGD--ELGEEDLDEDIEEADAAAEDVDVVID 72
KN L + + +E D+ DE GD EL + ++ +A + +
Sbjct 539 KNISLQLKQQSLLSEEEDYTTGSEVTEDEVGDEEELSRKQAGRKVKPKRSAKQTKKHI-- 596
Query 73 PNDPRLQRPEAHPSEGPRITTPFLTKFEKARIIGTRALQISMNA 116
+ P + + E + R ++PF ++ + +R+L+ + +A
Sbjct 597 -SSPSIHKKERQQEKASRDSSPFTVSLQRNKWDASRSLRFNQDA 639
> mmu:19649 Robo3, Rbig1, Rig-1, Rig1, Robo3a, Robo3b; roundabout
homolog 3 (Drosophila); K06755 roundabout, axon guidance
receptor 3
Length=1402
Score = 29.3 bits (64), Expect = 4.3, Method: Composition-based stats.
Identities = 14/52 (26%), Positives = 23/52 (44%), Gaps = 0/52 (0%)
Query 71 IDPNDPRLQRPEAHPSEGPRITTPFLTKFEKARIIGTRALQISMNAPLTIQL 122
+ D L RP P +G R + ++ ++G R LQ+S +QL
Sbjct 647 VQTQDSSLSRPAEDPWKGQRGLAEVAVRMQEPTVLGPRTLQVSWTVDGPVQL 698
> cel:T01B4.1 twk-21; TWiK family of potassium channels family
member (twk-21)
Length=522
Score = 29.3 bits (64), Expect = 4.8, Method: Composition-based stats.
Identities = 22/79 (27%), Positives = 35/79 (44%), Gaps = 13/79 (16%)
Query 30 ENDHMFAEPAFGDEFGDELGEEDLDEDIEEADAAAEDVDVVIDPNDPRLQRPEAHPSEGP 89
EN +M+A ++ ++L ++ DED ++AD ND + E P GP
Sbjct 314 ENQYMWALELIDQKYQEKLKQDMYDEDEKKADK-----------NDMHFSKKE--PVRGP 360
Query 90 RITTPFLTKFEKARIIGTR 108
RI L + +I G R
Sbjct 361 RILLQDLLRGPDLKISGGR 379
> tgo:TGME49_094740 armadillo/beta-catenin-like repeat-containing
protein
Length=3350
Score = 28.9 bits (63), Expect = 5.2, Method: Composition-based stats.
Identities = 16/39 (41%), Positives = 19/39 (48%), Gaps = 0/39 (0%)
Query 27 ADDENDHMFAEPAFGDEFGDELGEEDLDEDIEEADAAAE 65
A DE D EP FG+E G GE D++E D E
Sbjct 153 ARDEEDLDRKEPDFGEETGGRRGEGDIEETQRSQDEEKE 191
> mmu:19661 Rbp3, Irbp, MGC102323, Rbp-3; retinol binding protein
3, interstitial
Length=1234
Score = 28.9 bits (63), Expect = 5.9, Method: Composition-based stats.
Identities = 20/70 (28%), Positives = 32/70 (45%), Gaps = 3/70 (4%)
Query 48 LGEEDLDEDIEEA-DAAAEDVDVVIDPNDPR--LQRPEAHPSEGPRITTPFLTKFEKARI 104
+ EEDL + A +ED +++ PR RPE P+E P T T+ + R
Sbjct 363 VSEEDLVTKLNAGLQAVSEDPRLLVRATGPRDSSSRPETGPNESPAATPEVPTEEDARRA 422
Query 105 IGTRALQISM 114
+ Q+S+
Sbjct 423 LVDSVFQVSV 432
> pfa:PFA_0220w ubiquitin carboxyl-terminal hydrolase, putative
(EC:3.1.2.15)
Length=3499
Score = 28.5 bits (62), Expect = 6.8, Method: Composition-based stats.
Identities = 16/41 (39%), Positives = 22/41 (53%), Gaps = 0/41 (0%)
Query 7 QNKTQKNRKNTGLGFRVLKMADDENDHMFAEPAFGDEFGDE 47
Q + QK RKNT L V M D+ ++ F E F E+G +
Sbjct 3008 QMENQKIRKNTSLEKNVHHMNDNYDEINFTEKYFEQEYGSD 3048
> mmu:21885 Tle1, C230057C06Rik, Estm14, Grg1, Tle4l; transducin-like
enhancer of split 1, homolog of Drosophila E(spl)
Length=770
Score = 28.5 bits (62), Expect = 7.9, Method: Composition-based stats.
Identities = 14/51 (27%), Positives = 27/51 (52%), Gaps = 1/51 (1%)
Query 41 GDEFGDELGEEDLDE-DIEEADAAAEDVDVVIDPNDPRLQRPEAHPSEGPR 90
G EF ++ + +D+ D ++D D ++V+D ++ P A P+ PR
Sbjct 217 GPEFSSDIKKRKVDDKDNYDSDGDKSDDNLVVDVSNEDPSSPHASPTHSPR 267
> cpv:cgd7_5340 hypothetical protein
Length=548
Score = 28.1 bits (61), Expect = 9.6, Method: Composition-based stats.
Identities = 14/49 (28%), Positives = 26/49 (53%), Gaps = 0/49 (0%)
Query 30 ENDHMFAEPAFGDEFGDELGEEDLDEDIEEADAAAEDVDVVIDPNDPRL 78
E +F E A D++ + ED+ ++ D+ E +D V +P++P L
Sbjct 37 ERSQIFPEEANNDDYDNWKLPEDVSCNVWMMDSIGEFIDCVTNPDNPAL 85
> dre:562120 trpc2; transient receptor potential cation channel,
subfamily C, member 2; K04965 transient receptor potential
cation channel subfamily C member 2
Length=1140
Score = 28.1 bits (61), Expect = 9.6, Method: Compositional matrix adjust.
Identities = 13/32 (40%), Positives = 21/32 (65%), Gaps = 0/32 (0%)
Query 49 GEEDLDEDIEEADAAAEDVDVVIDPNDPRLQR 80
GE D+ D+EE++ A+ED +VV + N L +
Sbjct 937 GETDVSGDVEESNEASEDAEVVSEENKFELNQ 968
Lambda K H
0.313 0.134 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2807590228
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40