bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2571_orf1
Length=133
Score E
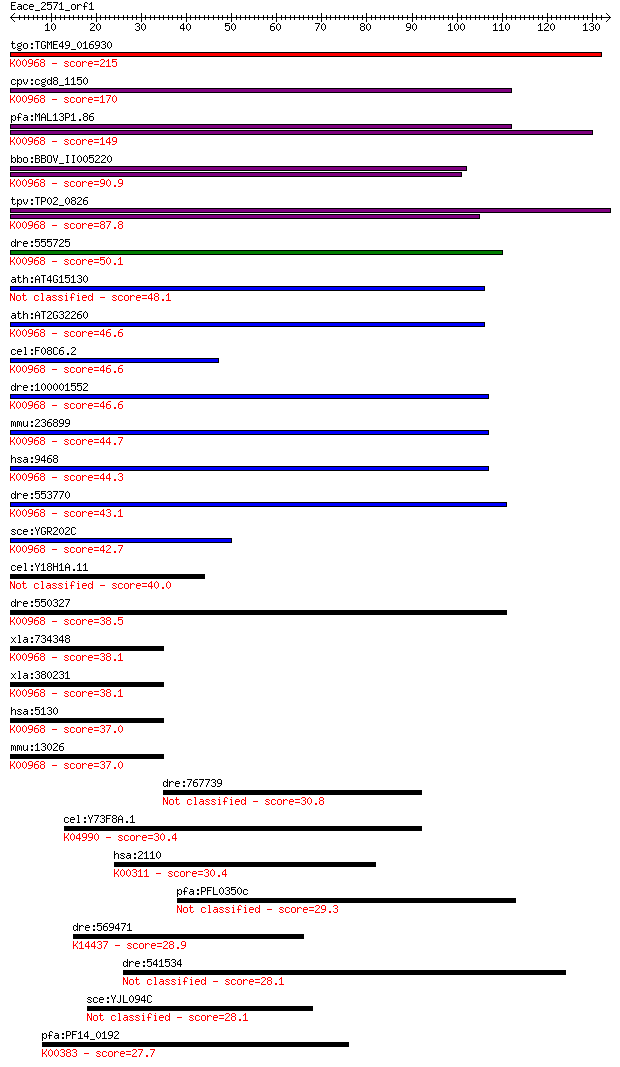
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_016930 cholinephosphate cytidylyltransferase, putat... 215 3e-56
cpv:cgd8_1150 choline-phosphate cytidylyltransferase ; K00968 ... 170 9e-43
pfa:MAL13P1.86 ctp; cholinephosphate cytidylyltransferase (EC:... 149 2e-36
bbo:BBOV_II005220 18.m06431; choline-phosphate cytidylyltransf... 90.9 9e-19
tpv:TP02_0826 cholinephosphate cytidylyltransferase; K00968 ch... 87.8 7e-18
dre:555725 pcyt1bb, MGC123291, zgc:123291; phosphate cytidylyl... 50.1 2e-06
ath:AT4G15130 CCT2; catalytic/ choline-phosphate cytidylyltran... 48.1 8e-06
ath:AT2G32260 CCT1; cholinephosphate cytidylyltransferase, put... 46.6 2e-05
cel:F08C6.2 hypothetical protein; K00968 choline-phosphate cyt... 46.6 2e-05
dre:100001552 pcyt1ba, pcyt1b; phosphate cytidylyltransferase ... 46.6 2e-05
mmu:236899 Pcyt1b, AW045697, CTTbeta; phosphate cytidylyltrans... 44.7 8e-05
hsa:9468 PCYT1B, CCTB, CTB; phosphate cytidylyltransferase 1, ... 44.3 1e-04
dre:553770 pcyt1ab, MGC110012, zgc:110012; phosphate cytidylyl... 43.1 2e-04
sce:YGR202C PCT1, BSR2, CCT1; Cholinephosphate cytidylyltransf... 42.7 3e-04
cel:Y18H1A.11 hypothetical protein 40.0 0.002
dre:550327 pcyt1aa, zgc:110237; phosphate cytidylyltransferase... 38.5 0.006
xla:734348 pcyt1a, MGC97881, ccta, cta, ctpct, pcyt1; phosphat... 38.1 0.007
xla:380231 pcyt1b, MGC53725, pcyt1a; phosphate cytidylyltransf... 38.1 0.007
hsa:5130 PCYT1A, CCTA, CT, CTA, CTPCT, PCYT1; phosphate cytidy... 37.0 0.017
mmu:13026 Pcyt1a, CTalpha, Cctalpha, Ctpct, Cttalpha; phosphat... 37.0 0.018
dre:767739 neurlb, MGC153175, zgc:153175; neuralized homolog b... 30.8 1.1
cel:Y73F8A.1 pkd-2; human PKD2 (polycystic kidney disease) rel... 30.4 1.6
hsa:2110 ETFDH, ETFQO, MADD; electron-transferring-flavoprotei... 30.4 1.6
pfa:PFL0350c conserved Plasmodium protein 29.3 3.1
dre:569471 chd7, KIAA0308, fd19h06, si:ch211-197o6.2, wu:cegs2... 28.9 4.5
dre:541534 vps33b, zgc:110803; vacuolar protein sorting 33B 28.1 7.1
sce:YJL094C KHA1; Kha1p 28.1 7.3
pfa:PF14_0192 glutathione reductase (EC:1.8.1.7); K00383 gluta... 27.7 9.4
> tgo:TGME49_016930 cholinephosphate cytidylyltransferase, putative
(EC:2.7.7.15); K00968 choline-phosphate cytidylyltransferase
[EC:2.7.7.15]
Length=329
Score = 215 bits (547), Expect = 3e-56, Method: Compositional matrix adjust.
Identities = 102/133 (76%), Positives = 113/133 (84%), Gaps = 2/133 (1%)
Query 1 STTDIIVRILQNYEDYVDRSLSRGVKPRDMNIGYTKANAIKMKKNMQRWGEKVSDELTKV 60
STTD+IVRILQNYEDYVDRSL RGV P+DMNIG+T ANAIKMKKNMQRWGEKVSDELTKV
Sbjct 196 STTDLIVRILQNYEDYVDRSLQRGVTPKDMNIGFTTANAIKMKKNMQRWGEKVSDELTKV 255
Query 61 TLTDRPLGVHFDESVEKLRNSIHQTYDNWRGRSHRLLRGFASKFEPMKSLIGIH--VNHH 118
TLTDRPLGV+FDESVEKLRNSIH+ YD+WR SH+ L+GFA FEPMKS IG+H H
Sbjct 256 TLTDRPLGVNFDESVEKLRNSIHEKYDSWRAHSHKFLKGFARTFEPMKSFIGLHHKSPRH 315
Query 119 QSSDDDAAISDGS 131
S ++D A SD S
Sbjct 316 TSDEEDGAASDAS 328
> cpv:cgd8_1150 choline-phosphate cytidylyltransferase ; K00968
choline-phosphate cytidylyltransferase [EC:2.7.7.15]
Length=341
Score = 170 bits (431), Expect = 9e-43, Method: Compositional matrix adjust.
Identities = 75/111 (67%), Positives = 94/111 (84%), Gaps = 0/111 (0%)
Query 1 STTDIIVRILQNYEDYVDRSLSRGVKPRDMNIGYTKANAIKMKKNMQRWGEKVSDELTKV 60
STTD++VRILQNYE+Y+DRSL RGV P ++NIGY KAN I+MKK +QRWGEKV++ELTKV
Sbjct 217 STTDLVVRILQNYEEYIDRSLQRGVTPDELNIGYMKANQIQMKKGIQRWGEKVTNELTKV 276
Query 61 TLTDRPLGVHFDESVEKLRNSIHQTYDNWRGRSHRLLRGFASKFEPMKSLI 111
TLTDRPLG+ FDESV+ +RN IH+++D WRG S + L GFA F+PM+SL
Sbjct 277 TLTDRPLGITFDESVDNIRNQIHKSFDAWRGVSKKYLEGFARTFDPMRSLF 327
> pfa:MAL13P1.86 ctp; cholinephosphate cytidylyltransferase (EC:2.7.7.15);
K00968 choline-phosphate cytidylyltransferase [EC:2.7.7.15]
Length=896
Score = 149 bits (376), Expect = 2e-36, Method: Compositional matrix adjust.
Identities = 66/111 (59%), Positives = 88/111 (79%), Gaps = 0/111 (0%)
Query 1 STTDIIVRILQNYEDYVDRSLSRGVKPRDMNIGYTKANAIKMKKNMQRWGEKVSDELTKV 60
STTD+IVRIL+NYEDY++RSL RG+ P ++NIG TKA +IKMKKN+ RWGEKV+DELTKV
Sbjct 175 STTDLIVRILKNYEDYIERSLQRGIHPNELNIGVTKAQSIKMKKNLIRWGEKVTDELTKV 234
Query 61 TLTDRPLGVHFDESVEKLRNSIHQTYDNWRGRSHRLLRGFASKFEPMKSLI 111
TLTD+PLG FD+ ++ +R+ +H + WR S +LL+ FA F+PM +I
Sbjct 235 TLTDKPLGTDFDQGIDIIRDKVHDLFKLWRYHSKKLLKDFAKSFDPMFIII 285
Score = 140 bits (354), Expect = 7e-34, Method: Compositional matrix adjust.
Identities = 68/130 (52%), Positives = 93/130 (71%), Gaps = 1/130 (0%)
Query 1 STTDIIVRILQNYEDYVDRSLSRGVKPRDMNIGYTKANAIKMKKNMQRWGEKVSDELTKV 60
STTD+IVRIL+NYEDY++RSL RG+ P ++NIG TKA +IKMKKN+ RWGEKV+DELTKV
Sbjct 760 STTDLIVRILKNYEDYIERSLQRGIHPNELNIGVTKAQSIKMKKNLIRWGEKVTDELTKV 819
Query 61 TLTDRPLGVHFDESVEKLRNSIHQTYDNWRGRSHRLLRGFASKFEPMKSLIGI-HVNHHQ 119
TLTD+PLG FD+ VE L+ + + W+ S++L+ F K E L I ++ ++
Sbjct 820 TLTDKPLGTDFDQGVENLQVKFKELFKIWKNASNKLITDFTRKLEATSYLTSIQNIIDYE 879
Query 120 SSDDDAAISD 129
+DD A S+
Sbjct 880 IENDDYASSN 889
> bbo:BBOV_II005220 18.m06431; choline-phosphate cytidylyltransferase
(EC:2.7.7.15); K00968 choline-phosphate cytidylyltransferase
[EC:2.7.7.15]
Length=546
Score = 90.9 bits (224), Expect = 9e-19, Method: Compositional matrix adjust.
Identities = 42/101 (41%), Positives = 68/101 (67%), Gaps = 3/101 (2%)
Query 1 STTDIIVRILQNYEDYVDRSLSRGVKPRDMNIGYTKANAIKMKKNMQRWGEKVSDELTKV 60
TTD++VRILQ+YEDY+++S++ GV P+D+NIG T AN+I++KK + +W +++D++T
Sbjct 141 CTTDLVVRILQHYEDYIEKSINTGVNPKDLNIGTTMANSIRVKKKINKWVRQITDQITS- 199
Query 61 TLTDRPLGVHFDESVEKLRNSIHQTYDNWRGRSHRLLRGFA 101
TD LG + +E++R I + + W R LR FA
Sbjct 200 --TDVSLGNKLEGKIEEIRAGIATSLEQWIERYIHALRDFA 238
Score = 90.9 bits (224), Expect = 9e-19, Method: Compositional matrix adjust.
Identities = 44/100 (44%), Positives = 66/100 (66%), Gaps = 4/100 (4%)
Query 1 STTDIIVRILQNYEDYVDRSLSRGVKPRDMNIGYTKANAIKMKKNMQRWGEKVSDELTKV 60
ST++I++RIL+NYE Y+ RSL RGV D+ IG+T AN+IK+K +++ W +K + E+ K
Sbjct 441 STSNIMMRILRNYETYIKRSLERGVGREDLKIGFTTANSIKLKSSIENWQKKFTQEVHKA 500
Query 61 TLTDRPLGVHFDESVEKLRNSIHQTYDNWRGRSHRLLRGF 100
TLTD P+G FD ++K I + D WR S ++ F
Sbjct 501 TLTDHPVGHEFDRLIDK----IVEVVDGWRKDSKVMIENF 536
> tpv:TP02_0826 cholinephosphate cytidylyltransferase; K00968
choline-phosphate cytidylyltransferase [EC:2.7.7.15]
Length=523
Score = 87.8 bits (216), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 45/148 (30%), Positives = 82/148 (55%), Gaps = 15/148 (10%)
Query 1 STTDIIVRILQNYEDYVDRSLSRGVKPRDMNIGYTKANAIKMKKNMQRWGEKVSDELTKV 60
STT+ I++ILQNYED++D+ L G+KP D+NI T +I +KK + +K+S ++T++
Sbjct 143 STTECIIKILQNYEDFIDKFLKNGLKPSDLNIPITTGKSILLKKTILALVDKLSTQITQL 202
Query 61 TLTDRPLGVHFDESVEKLRNSIHQTYDNWRGRSHRLLRGF---------------ASKFE 105
TLTD PLG F++ ++ +R+S+ + W + +L F + E
Sbjct 203 TLTDEPLGAGFNDRIDLVRSSLFKAIAIWLNKHKIVLEDFKEIPYVDPAETVYSKSVTME 262
Query 106 PMKSLIGIHVNHHQSSDDDAAISDGSGL 133
P++S+ +++ DD+ + G+
Sbjct 263 PVESIPKLNLPSGNLKDDEEVVVYTYGV 290
Score = 68.2 bits (165), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 33/107 (30%), Positives = 64/107 (59%), Gaps = 3/107 (2%)
Query 1 STTDIIVRILQNYEDYVDRSLSRGVKPRDMNIGYTKANAIKMKKNMQRWGEKVSDELTKV 60
TT +++ IL+NY+ Y+ RSL RGV R++ +GY K ++K+K +++ + +K++DE+ ++
Sbjct 406 CTTSLMLNILKNYDLYLLRSLDRGVCRRELKLGYAKERSLKVKYSIKNFQKKLNDEIIQL 465
Query 61 TLTDR---PLGVHFDESVEKLRNSIHQTYDNWRGRSHRLLRGFASKF 104
TL D+ G FD++V + + +D WR + F + +
Sbjct 466 TLMDKHIVSFGHKFDKNVNVMVTKMQNAFDAWRRDYINFISEFVNYY 512
> dre:555725 pcyt1bb, MGC123291, zgc:123291; phosphate cytidylyltransferase
1, choline, beta b; K00968 choline-phosphate cytidylyltransferase
[EC:2.7.7.15]
Length=299
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 30/109 (27%), Positives = 51/109 (46%), Gaps = 11/109 (10%)
Query 1 STTDIIVRILQNYEDYVDRSLSRGVKPRDMNIGYTKANAIKMKKNMQRWGEKVSDELTKV 60
ST+DII RI+++Y+ YV R+L RG R++N+G+ K ++++ + R E V
Sbjct 196 STSDIITRIVRDYDVYVRRNLQRGYTARELNVGFIKEKKYRLQQQVDRMKETV------- 248
Query 61 TLTDRPLGVHFDESVEKLRNSIHQTYDNWRGRSHRLLRGFASKFEPMKS 109
R + V ++ H W +S + F F P K+
Sbjct 249 ----RTVEEKSKHLVHRVEEKSHDLIYKWEEKSREFIGNFLELFGPDKA 293
> ath:AT4G15130 CCT2; catalytic/ choline-phosphate cytidylyltransferase/
nucleotidyltransferase (EC:2.7.7.15)
Length=304
Score = 48.1 bits (113), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 28/105 (26%), Positives = 52/105 (49%), Gaps = 16/105 (15%)
Query 1 STTDIIVRILQNYEDYVDRSLSRGVKPRDMNIGYTKANAIKMKKNMQRWGEKVSDELTKV 60
ST+DII+RI+++Y YV R+L RG ++ + + + +++ +++ EKV ++ K+
Sbjct 147 STSDIIMRIVKDYNQYVLRNLDRGYSREELGVSFEEKR-LRVNMRLKKLQEKVKEQQEKI 205
Query 61 TLTDRPLGVHFDESVEKLRNSIHQTYDNWRGRSHRLLRGFASKFE 105
+ G+H DE W + R + GF FE
Sbjct 206 QTVAKTAGMHHDE---------------WLENADRWVAGFLEMFE 235
> ath:AT2G32260 CCT1; cholinephosphate cytidylyltransferase, putative
/ phosphorylcholine transferase, putative / CTP:phosphocholine
cytidylyltransferase, putative (EC:2.7.7.15); K00968
choline-phosphate cytidylyltransferase [EC:2.7.7.15]
Length=332
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 30/105 (28%), Positives = 54/105 (51%), Gaps = 15/105 (14%)
Query 1 STTDIIVRILQNYEDYVDRSLSRGVKPRDMNIGYTKANAIKMKKNMQRWGEKVSDELTKV 60
ST+DII+RI+++Y YV R+L RG D+ + + K +++ +++ E+V ++ +V
Sbjct 162 STSDIIMRIVKDYNQYVMRNLDRGYSREDLGVSFVKEKRLRVNMRLKKLQERVKEQQERV 221
Query 61 TLTDRPLGVHFDESVEKLRNSIHQTYDNWRGRSHRLLRGFASKFE 105
G ++V+ LRN W + R + GF FE
Sbjct 222 -------GEKI-QTVKMLRNE-------WVENADRWVAGFLEIFE 251
> cel:F08C6.2 hypothetical protein; K00968 choline-phosphate cytidylyltransferase
[EC:2.7.7.15]
Length=362
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 16/46 (34%), Positives = 34/46 (73%), Gaps = 0/46 (0%)
Query 1 STTDIIVRILQNYEDYVDRSLSRGVKPRDMNIGYTKANAIKMKKNM 46
ST+D++ RI+++Y+ YV R+L RG P+++N+G+ A+ +++ +
Sbjct 205 STSDVVCRIIRDYDKYVRRNLQRGYSPKELNVGFLAASKYQIQNKV 250
> dre:100001552 pcyt1ba, pcyt1b; phosphate cytidylyltransferase
1, choline, beta a (EC:2.7.7.15); K00968 choline-phosphate
cytidylyltransferase [EC:2.7.7.15]
Length=343
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 28/106 (26%), Positives = 50/106 (47%), Gaps = 11/106 (10%)
Query 1 STTDIIVRILQNYEDYVDRSLSRGVKPRDMNIGYTKANAIKMKKNMQRWGEKVSDELTKV 60
ST+D+I RI+++Y+ Y R+L RG +++N+ Y +++ + R EKV
Sbjct 175 STSDLITRIVRDYDVYARRNLQRGYTAKELNVSYINEKKYRLQNQVDRMKEKVR------ 228
Query 61 TLTDRPLGVHFDESVEKLRNSIHQTYDNWRGRSHRLLRGFASKFEP 106
T+ ++ HF V ++ H W +S + F F P
Sbjct 229 TVEEK--SKHF---VYRVEEKSHDLIQKWEEKSREFIGNFLELFGP 269
> mmu:236899 Pcyt1b, AW045697, CTTbeta; phosphate cytidylyltransferase
1, choline, beta isoform (EC:2.7.7.15); K00968 choline-phosphate
cytidylyltransferase [EC:2.7.7.15]
Length=339
Score = 44.7 bits (104), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 27/108 (25%), Positives = 48/108 (44%), Gaps = 15/108 (13%)
Query 1 STTDIIVRILQNYEDYVDRSLSRGVKPRDMNIGYTKANAIKMKKNMQRWGEKVS--DELT 58
ST+DII RI+++Y+ Y R+L RG +++N+ + + + + + EKV +E +
Sbjct 171 STSDIITRIVRDYDVYARRNLQRGYTAKELNVSFINEKKYRFQNQVDKMKEKVKNVEERS 230
Query 59 KVTLTDRPLGVHFDESVEKLRNSIHQTYDNWRGRSHRLLRGFASKFEP 106
K E V ++ H W +S + F F P
Sbjct 231 K-------------EFVNRVEEKSHDLIQKWEEKSREFIGNFLELFGP 265
> hsa:9468 PCYT1B, CCTB, CTB; phosphate cytidylyltransferase 1,
choline, beta (EC:2.7.7.15); K00968 choline-phosphate cytidylyltransferase
[EC:2.7.7.15]
Length=351
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 27/108 (25%), Positives = 48/108 (44%), Gaps = 15/108 (13%)
Query 1 STTDIIVRILQNYEDYVDRSLSRGVKPRDMNIGYTKANAIKMKKNMQRWGEKVS--DELT 58
ST+DII RI+++Y+ Y R+L RG +++N+ + + + + + EKV +E +
Sbjct 183 STSDIITRIVRDYDVYARRNLQRGYTAKELNVSFINEKRYRFQNQVDKMKEKVKNVEERS 242
Query 59 KVTLTDRPLGVHFDESVEKLRNSIHQTYDNWRGRSHRLLRGFASKFEP 106
K E V ++ H W +S + F F P
Sbjct 243 K-------------EFVNRVEEKSHDLIQKWEEKSREFIGNFLELFGP 277
> dre:553770 pcyt1ab, MGC110012, zgc:110012; phosphate cytidylyltransferase
1, choline, alpha b; K00968 choline-phosphate
cytidylyltransferase [EC:2.7.7.15]
Length=359
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 27/110 (24%), Positives = 51/110 (46%), Gaps = 11/110 (10%)
Query 1 STTDIIVRILQNYEDYVDRSLSRGVKPRDMNIGYTKANAIKMKKNMQRWGEKVSDELTKV 60
ST+DII RI+++Y+ YV R+L RG +++N+ + +++++ + +KV + K
Sbjct 198 STSDIITRIVRDYDVYVRRNLQRGYTAKELNVSFINEKKYHLQEHVDKVKQKVRNVEEKS 257
Query 61 TLTDRPLGVHFDESVEKLRNSIHQTYDNWRGRSHRLLRGFASKFEPMKSL 110
E V+K+ W +S + F F P +L
Sbjct 258 K-----------EFVQKVEEKGIDLIQKWEEKSREFIGNFLQMFGPDGAL 296
> sce:YGR202C PCT1, BSR2, CCT1; Cholinephosphate cytidylyltransferase,
also known as CTP:phosphocholine cytidylyltransferase,
rate-determining enzyme of the CDP-choline pathway for phosphatidylcholine
synthesis, inhibited by Sec14p, activated
upon lipid-binding (EC:2.7.7.15); K00968 choline-phosphate
cytidylyltransferase [EC:2.7.7.15]
Length=424
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 16/49 (32%), Positives = 35/49 (71%), Gaps = 0/49 (0%)
Query 1 STTDIIVRILQNYEDYVDRSLSRGVKPRDMNIGYTKANAIKMKKNMQRW 49
ST+DII +I+++Y+ Y+ R+ +RG +++N+ + K N ++ KK++ +
Sbjct 228 STSDIITKIIRDYDKYLMRNFARGATRQELNVSWLKKNELEFKKHINEF 276
> cel:Y18H1A.11 hypothetical protein
Length=272
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 16/43 (37%), Positives = 29/43 (67%), Gaps = 0/43 (0%)
Query 1 STTDIIVRILQNYEDYVDRSLSRGVKPRDMNIGYTKANAIKMK 43
ST+D I RI+++Y+ YV R+L RG D+N+G+ + +++
Sbjct 171 STSDSICRIIRDYDTYVRRNLQRGYSATDLNVGFFTTSKYRLQ 213
> dre:550327 pcyt1aa, zgc:110237; phosphate cytidylyltransferase
1, choline, alpha a (EC:2.7.7.15); K00968 choline-phosphate
cytidylyltransferase [EC:2.7.7.15]
Length=374
Score = 38.5 bits (88), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 30/110 (27%), Positives = 52/110 (47%), Gaps = 11/110 (10%)
Query 1 STTDIIVRILQNYEDYVDRSLSRGVKPRDMNIGYTKANAIKMKKNMQRWGEKVSDELTKV 60
ST+DII RI+++Y+ YV R+L RG +++N+ + + K N+Q +KV ++ V
Sbjct 198 STSDIITRIVRDYDVYVRRNLQRGYTAKELNVSFIN----EKKYNLQERVDKVKKKVKDV 253
Query 61 TLTDRPLGVHFDESVEKLRNSIHQTYDNWRGRSHRLLRGFASKFEPMKSL 110
+ E V+K+ W +S + F F P +L
Sbjct 254 EEKSK-------EFVQKVEEKSIDLIQKWEEKSREFIGNFLQMFGPEGAL 296
> xla:734348 pcyt1a, MGC97881, ccta, cta, ctpct, pcyt1; phosphate
cytidylyltransferase 1, choline, alpha (EC:2.7.7.15); K00968
choline-phosphate cytidylyltransferase [EC:2.7.7.15]
Length=367
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 15/34 (44%), Positives = 26/34 (76%), Gaps = 0/34 (0%)
Query 1 STTDIIVRILQNYEDYVDRSLSRGVKPRDMNIGY 34
ST+DII RI+++Y+ YV R+L RG +++N+ +
Sbjct 201 STSDIITRIVRDYDVYVRRNLQRGYTAKELNVSF 234
> xla:380231 pcyt1b, MGC53725, pcyt1a; phosphate cytidylyltransferase
1, choline, beta (EC:2.7.7.15); K00968 choline-phosphate
cytidylyltransferase [EC:2.7.7.15]
Length=366
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 15/34 (44%), Positives = 26/34 (76%), Gaps = 0/34 (0%)
Query 1 STTDIIVRILQNYEDYVDRSLSRGVKPRDMNIGY 34
ST+DII RI+++Y+ YV R+L RG +++N+ +
Sbjct 201 STSDIITRIVRDYDVYVRRNLQRGYTAKELNVSF 234
> hsa:5130 PCYT1A, CCTA, CT, CTA, CTPCT, PCYT1; phosphate cytidylyltransferase
1, choline, alpha (EC:2.7.7.15); K00968 choline-phosphate
cytidylyltransferase [EC:2.7.7.15]
Length=367
Score = 37.0 bits (84), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 14/34 (41%), Positives = 25/34 (73%), Gaps = 0/34 (0%)
Query 1 STTDIIVRILQNYEDYVDRSLSRGVKPRDMNIGY 34
ST+DII RI+++Y+ Y R+L RG +++N+ +
Sbjct 201 STSDIITRIVRDYDVYARRNLQRGYTAKELNVSF 234
> mmu:13026 Pcyt1a, CTalpha, Cctalpha, Ctpct, Cttalpha; phosphate
cytidylyltransferase 1, choline, alpha isoform (EC:2.7.7.15);
K00968 choline-phosphate cytidylyltransferase [EC:2.7.7.15]
Length=367
Score = 37.0 bits (84), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 14/34 (41%), Positives = 25/34 (73%), Gaps = 0/34 (0%)
Query 1 STTDIIVRILQNYEDYVDRSLSRGVKPRDMNIGY 34
ST+DII RI+++Y+ Y R+L RG +++N+ +
Sbjct 201 STSDIITRIVRDYDVYARRNLQRGYTAKELNVSF 234
> dre:767739 neurlb, MGC153175, zgc:153175; neuralized homolog
b (Drosophila)
Length=498
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 19/57 (33%), Positives = 29/57 (50%), Gaps = 6/57 (10%)
Query 35 TKANAIKMKKNMQRWGEKVSDELTKVTLTDRPLGVHFDESVEKLRNSIHQTYDNWRG 91
TK + I M K QR +++ +T T RP+GV+ E++R I +T W G
Sbjct 17 TKGSQIVMDKT-QRSVRRIASFCNAITFTSRPVGVY-----EQVRLKITKTQGCWSG 67
> cel:Y73F8A.1 pkd-2; human PKD2 (polycystic kidney disease) related
family member (pkd-2); K04990 polycystin 2L1
Length=716
Score = 30.4 bits (67), Expect = 1.6, Method: Composition-based stats.
Identities = 19/86 (22%), Positives = 40/86 (46%), Gaps = 7/86 (8%)
Query 13 YEDYVDRSLSRGVKPRDMNIGYTKANAIKMKKNM-QRWGEKVSDELTKVTLTDRPLG--- 68
YEDY G +D+N +T+ N M +++ ++ E ++DE+ ++T R
Sbjct 578 YEDYKLMLYRAGYAEKDINEAFTRFNVTSMTEHVPEKVAEDIADEVARMTEQKRNYMENH 637
Query 69 ---VHFDESVEKLRNSIHQTYDNWRG 91
+ + V++++ S+ D G
Sbjct 638 RDYANLNRRVDQMQESVFSIVDRIEG 663
> hsa:2110 ETFDH, ETFQO, MADD; electron-transferring-flavoprotein
dehydrogenase (EC:1.5.5.1); K00311 electron-transferring-flavoprotein
dehydrogenase [EC:1.5.5.1]
Length=617
Score = 30.4 bits (67), Expect = 1.6, Method: Composition-based stats.
Identities = 17/58 (29%), Positives = 27/58 (46%), Gaps = 1/58 (1%)
Query 24 GVKPRDMNIGYTKANAIKMKKNMQRWGEKVSDELTKVTLTDRPLGVHFDESVEKLRNS 81
G P MN+ K MK + E + ++LT L + +G+H E + L+NS
Sbjct 386 GCSPGFMNVPKIKGTHTAMKSGILA-AESIFNQLTSENLQSKTIGLHVTEYEDNLKNS 442
> pfa:PFL0350c conserved Plasmodium protein
Length=2612
Score = 29.3 bits (64), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 20/76 (26%), Positives = 39/76 (51%), Gaps = 4/76 (5%)
Query 38 NAIKMKKNMQRWGEKVSDELTKVTLTDRPLGVHFDESVEKLRNSIHQTY-DNWRGRSHRL 96
N KM KN+++ EK+ DE+ K R + + +E ++ + + Q Y +N+ L
Sbjct 1162 NKEKMIKNVEKENEKLKDEIEKER---RNMIQNLEEEKKEFKLYLEQKYKENFENEKSGL 1218
Query 97 LRGFASKFEPMKSLIG 112
+ F + E +++ IG
Sbjct 1219 AKKFDEENEKLQNEIG 1234
> dre:569471 chd7, KIAA0308, fd19h06, si:ch211-197o6.2, wu:cegs2051,
wu:fb37f10, wu:fb39h04, wu:fd19h06; chromodomain helicase
DNA binding protein 7; K14437 chromodomain-helicase-DNA-binding
protein 7 [EC:3.6.4.12]
Length=3094
Score = 28.9 bits (63), Expect = 4.5, Method: Composition-based stats.
Identities = 15/56 (26%), Positives = 29/56 (51%), Gaps = 5/56 (8%)
Query 15 DYVDRSLSRGVKPRDMNIGYTKANAIKMKKNM-----QRWGEKVSDELTKVTLTDR 65
D D+ + + +P+D GY ++ +++KN+ RWG+ +S K L +R
Sbjct 1634 DSEDKPIQKPRRPQDRTQGYPRSECFRVEKNLLVYGWGRWGDILSHGRFKRPLRER 1689
> dre:541534 vps33b, zgc:110803; vacuolar protein sorting 33B
Length=617
Score = 28.1 bits (61), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 23/98 (23%), Positives = 39/98 (39%), Gaps = 6/98 (6%)
Query 26 KPRDMNIGYTKANAIKMKKNMQRWGEKVSDELTKVTLTDRPLGVHFDESVEKLRNSIHQT 85
K +++ Y K + ++Q+ V+DEL + R L +H S ++ Q
Sbjct 302 KAKNLQTAYDKRRGM----DIQQMKAFVADELKGLKQEHRLLSLHIGASESIMKKKTKQD 357
Query 86 YDNWRGRSHRLLRGFASKFEPMKSLIGIHVNHHQSSDD 123
+ H LL GF + + I H+N S D
Sbjct 358 FQELLKTEHSLLEGF--EIRECIAYIEEHINRQVSMID 393
> sce:YJL094C KHA1; Kha1p
Length=873
Score = 28.1 bits (61), Expect = 7.3, Method: Composition-based stats.
Identities = 16/50 (32%), Positives = 25/50 (50%), Gaps = 3/50 (6%)
Query 18 DRSLSRGVKPRDMNIGYTKANAIKMKKNMQRWGEKVSDELTKVTLTDRPL 67
+ SLSR D + K+N K+KK + W + V D T +++ D L
Sbjct 517 ETSLSRMTTATDSTL---KSNTFKIKKMVHIWSKSVDDVDTNLSVIDEKL 563
> pfa:PF14_0192 glutathione reductase (EC:1.8.1.7); K00383 glutathione
reductase (NADPH) [EC:1.8.1.7]
Length=500
Score = 27.7 bits (60), Expect = 9.4, Method: Composition-based stats.
Identities = 20/68 (29%), Positives = 36/68 (52%), Gaps = 9/68 (13%)
Query 8 RILQNYEDYVDRSLSRGVKPRDMNIGYTKANAIKMKKNMQRWGEKVSDELTKVTLTDRPL 67
RIL+ +++ V L +K ++NI T A+ +++KK VSD+ + L+D +
Sbjct 210 RILRKFDESVINVLENDMKKNNINI-VTFADVVEIKK--------VSDKNLSIHLSDGRI 260
Query 68 GVHFDESV 75
HFD +
Sbjct 261 YEHFDHVI 268
Lambda K H
0.316 0.133 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2165519004
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40