bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2527_orf1
Length=127
Score E
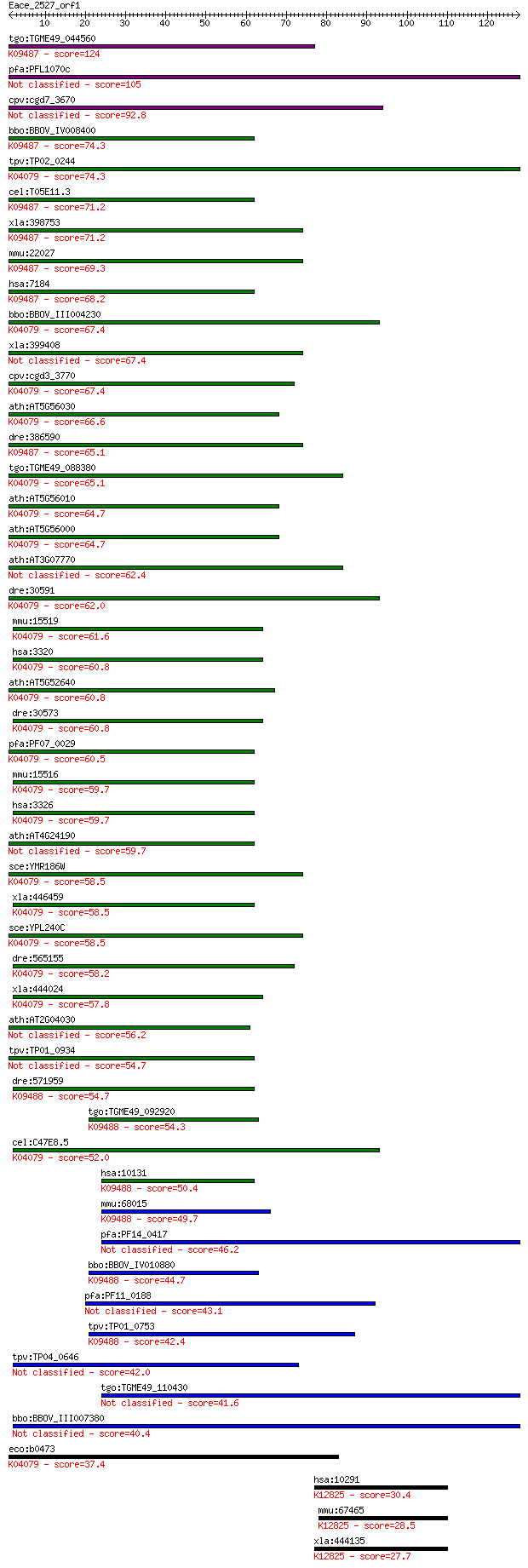
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_044560 heat shock protein 90, putative (EC:2.7.13.3... 124 6e-29
pfa:PFL1070c endoplasmin homolog precursor, putative 105 3e-23
cpv:cgd7_3670 heat shock protein 90 (Hsp90), signal peptide pl... 92.8 2e-19
bbo:BBOV_IV008400 23.m06066; heat shock protein 90; K09487 hea... 74.3 9e-14
tpv:TP02_0244 heat shock protein 90; K04079 molecular chaperon... 74.3 1e-13
cel:T05E11.3 hypothetical protein; K09487 heat shock protein 9... 71.2 7e-13
xla:398753 hypothetical protein MGC68448; K09487 heat shock pr... 71.2 8e-13
mmu:22027 Hsp90b1, ERp99, GRP94, TA-3, Targ2, Tra-1, Tra1, end... 69.3 2e-12
hsa:7184 HSP90B1, ECGP, GP96, GRP94, TRA1; heat shock protein ... 68.2 6e-12
bbo:BBOV_III004230 17.m07381; hsp90 protein; K04079 molecular ... 67.4 9e-12
xla:399408 hsp90b1, ecgp, gp96, grp94, tra1; heat shock protei... 67.4 1e-11
cpv:cgd3_3770 Hsp90 ; K04079 molecular chaperone HtpG 67.4 1e-11
ath:AT5G56030 HSP81-2 (HEAT SHOCK PROTEIN 81-2); ATP binding; ... 66.6 2e-11
dre:386590 hsp90b1, GP96, GRP94, fb61d09, tra-1, tra1, wu:fb61... 65.1 5e-11
tgo:TGME49_088380 heat shock protein 90 ; K04079 molecular cha... 65.1 5e-11
ath:AT5G56010 HSP81-3; ATP binding / unfolded protein binding;... 64.7 7e-11
ath:AT5G56000 heat shock protein 81-4 (HSP81-4); K04079 molecu... 64.7 8e-11
ath:AT3G07770 ATP binding 62.4 4e-10
dre:30591 hsp90a.1, fb17b01, hsp90, hsp90a, hsp90alpha, wu:fb1... 62.0 4e-10
mmu:15519 Hsp90aa1, 86kDa, 89kDa, AL024080, AL024147, Hsp86-1,... 61.6 7e-10
hsa:3320 HSP90AA1, FLJ31884, HSP86, HSP89A, HSP90A, HSP90N, HS... 60.8 1e-09
ath:AT5G52640 ATHSP90.1 (HEAT SHOCK PROTEIN 90.1); ATP binding... 60.8 1e-09
dre:30573 hsp90ab1, hsp90b, hsp90beta, wu:fa29f01, wu:fa91e11,... 60.8 1e-09
pfa:PF07_0029 heat shock protein 86; K04079 molecular chaperon... 60.5 1e-09
mmu:15516 Hsp90ab1, 90kDa, AL022974, C81438, Hsp84, Hsp84-1, H... 59.7 2e-09
hsa:3326 HSP90AB1, D6S182, FLJ26984, HSP90-BETA, HSP90B, HSPC2... 59.7 2e-09
ath:AT4G24190 SHD; SHD (SHEPHERD); ATP binding / unfolded prot... 59.7 2e-09
sce:YMR186W HSC82, HSP90; Cytoplasmic chaperone of the Hsp90 f... 58.5 5e-09
xla:446459 hsp90ab1, hsp90b, hsp90beta; heat shock protein 90k... 58.5 5e-09
sce:YPL240C HSP82, HSP90; Hsp82p; K04079 molecular chaperone HtpG 58.5 5e-09
dre:565155 hsp90a.2, cb820, hsp90a2; heat shock protein 90-alp... 58.2 7e-09
xla:444024 hsp90aa1.1, MGC82579, hsp86, hsp89, hsp90, hsp90a, ... 57.8 9e-09
ath:AT2G04030 CR88; CR88; ATP binding 56.2 3e-08
tpv:TP01_0934 heat shock protein 90 54.7
dre:571959 trap1, fc85a11, wu:fc85a11; TNF receptor-associated... 54.7 7e-08
tgo:TGME49_092920 heat shock protein 90, putative ; K09488 TNF... 54.3 1e-07
cel:C47E8.5 daf-21; abnormal DAuer Formation family member (da... 52.0 5e-07
hsa:10131 TRAP1, HSP75, HSP90L; TNF receptor-associated protei... 50.4 1e-06
mmu:68015 Trap1, 2410002K23Rik, HSP75; TNF receptor-associated... 49.7 3e-06
pfa:PF14_0417 HSP90 46.2 3e-05
bbo:BBOV_IV010880 23.m05763; heat shock protein 75; K09488 TNF... 44.7 7e-05
pfa:PF11_0188 heat shock protein 90, putative 43.1 2e-04
tpv:TP01_0753 heat shock protein 75; K09488 TNF receptor-assoc... 42.4 4e-04
tpv:TP04_0646 heat shock protein 90 42.0
tgo:TGME49_110430 heat shock protein 90, putative (EC:3.2.1.3 ... 41.6 6e-04
bbo:BBOV_III007380 17.m07646; heat shock protein 90 40.4
eco:b0473 htpG, ECK0467, JW0462; molecular chaperone HSP90 fam... 37.4 0.013
hsa:10291 SF3A1, PRP21, PRPF21, SAP114, SF3A120; splicing fact... 30.4 1.3
mmu:67465 Sf3a1, 1200014H24Rik, 5930416L09Rik, AI159724; splic... 28.5 5.2
xla:444135 MGC80562 protein; K12825 splicing factor 3A subunit 1 27.7
> tgo:TGME49_044560 heat shock protein 90, putative (EC:2.7.13.3);
K09487 heat shock protein 90kDa beta
Length=847
Score = 124 bits (311), Expect = 6e-29, Method: Compositional matrix adjust.
Identities = 53/76 (69%), Positives = 67/76 (88%), Gaps = 0/76 (0%)
Query 1 VWESSADAKYYIDKDPRGNTLERGTCVVLHLKEDATEFASVDRLRELATRYSQFMSYPIY 60
+WESSADAK+++ KDPRGNTL RGTCV LHLKEDATEF + +L++L TR+SQFMSYPIY
Sbjct 229 IWESSADAKFHVAKDPRGNTLGRGTCVTLHLKEDATEFLNEWKLKDLTTRFSQFMSYPIY 288
Query 61 LKVKQTVSEEVPVEDE 76
++ +TV+EEVP+EDE
Sbjct 289 VRTSRTVTEEVPIEDE 304
> pfa:PFL1070c endoplasmin homolog precursor, putative
Length=821
Score = 105 bits (262), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 60/127 (47%), Positives = 84/127 (66%), Gaps = 11/127 (8%)
Query 1 VWESSADAKYYIDKDPRGNTLERGTCVVLHLKEDATEFASVDRLRELATRYSQFMSYPIY 60
+WES+ADAK+ I KDPRG TL+RGT + LHLKEDAT + +L +L ++YSQF+ +PIY
Sbjct 217 IWESTADAKFTIYKDPRGATLKRGTRISLHLKEDATNLLNDKKLMDLISKYSQFIQFPIY 276
Query 61 LKVKQTVSEEVPVEDEKEDKAEDKEKSEEEDVQVQDVDEETDDKEAKKKKTKTVEREVER 120
L + +EEV + D +D D + V+V EETDD KKT+TVE++V++
Sbjct 277 LLHENVYTEEV-LADIAKDMVND---PNYDSVKV----EETDDPN---KKTRTVEKKVKK 325
Query 121 WDRINEQ 127
W +NEQ
Sbjct 326 WTLMNEQ 332
> cpv:cgd7_3670 heat shock protein 90 (Hsp90), signal peptide
plus ER retention motif
Length=787
Score = 92.8 bits (229), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 44/93 (47%), Positives = 66/93 (70%), Gaps = 2/93 (2%)
Query 1 VWESSADAKYYIDKDPRGNTLERGTCVVLHLKEDATEFASVDRLRELATRYSQFMSYPIY 60
VWESSAD + + DPRGNT++RGT +VL LKEDATEF + +L++L RYSQF+++PIY
Sbjct 230 VWESSADGSFRVSLDPRGNTIKRGTTIVLSLKEDATEFMNFSKLKDLVLRYSQFINFPIY 289
Query 61 LKVKQTVSEEVPVEDEKEDKAEDKEKSEEEDVQ 93
+ + V++ +DE +K E K + E+ +V+
Sbjct 290 IYNPEGVNKS--EKDESGEKKESKGRWEQVNVE 320
> bbo:BBOV_IV008400 23.m06066; heat shock protein 90; K09487 heat
shock protein 90kDa beta
Length=795
Score = 74.3 bits (181), Expect = 9e-14, Method: Compositional matrix adjust.
Identities = 33/62 (53%), Positives = 48/62 (77%), Gaps = 1/62 (1%)
Query 1 VWESSADAKYYIDKDPRGNTL-ERGTCVVLHLKEDATEFASVDRLRELATRYSQFMSYPI 59
VW+SSAD KY + +DP+GNTL E GT + L L+EDATE+ +D++ EL ++SQF+ +PI
Sbjct 232 VWKSSADTKYELYEDPKGNTLGEHGTQITLFLREDATEYLEIDKIEELIKKHSQFVRFPI 291
Query 60 YL 61
Y+
Sbjct 292 YV 293
> tpv:TP02_0244 heat shock protein 90; K04079 molecular chaperone
HtpG
Length=721
Score = 74.3 bits (181), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 56/137 (40%), Positives = 83/137 (60%), Gaps = 12/137 (8%)
Query 1 VWESSADAKYYIDKDPRGNTLERGTCVVLHLKEDATEFASVDRLRELATRYSQFMSYPIY 60
VWES+A + + KD L+RGT ++LHLKED TE+ RL+EL ++S+F+S+PI
Sbjct 154 VWESTASGHFTVKKDDSHEPLKRGTRLILHLKEDQTEYLEERRLKELVKKHSEFISFPIS 213
Query 61 LKVKQTVSEEVPVEDEKEDKAEDKEKSEE-------EDV---QVQDVDEETDDKEAKKKK 110
L V++T +E V D++ + EDK+ EE EDV +V DV +E + KE KKKK
Sbjct 214 LSVEKT--QETEVTDDEAELDEDKKPEEEKPKDDKVEDVTDEKVTDVTDEEEKKEEKKKK 271
Query 111 TKTVEREVERWDRINEQ 127
+ V W+ +N+Q
Sbjct 272 KRKVTNVTREWEMLNKQ 288
> cel:T05E11.3 hypothetical protein; K09487 heat shock protein
90kDa beta
Length=760
Score = 71.2 bits (173), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 31/61 (50%), Positives = 45/61 (73%), Gaps = 1/61 (1%)
Query 1 VWESSADAKYYIDKDPRGNTLERGTCVVLHLKEDATEFASVDRLRELATRYSQFMSYPIY 60
+WES + A + I KDPRGNTL+RGT + L+LKE+A +F D L+ L +YSQF+++ I+
Sbjct 210 IWESDS-ASFTISKDPRGNTLKRGTQITLYLKEEAADFLEPDTLKNLVHKYSQFINFDIF 268
Query 61 L 61
L
Sbjct 269 L 269
> xla:398753 hypothetical protein MGC68448; K09487 heat shock
protein 90kDa beta
Length=805
Score = 71.2 bits (173), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 30/73 (41%), Positives = 53/73 (72%), Gaps = 1/73 (1%)
Query 1 VWESSADAKYYIDKDPRGNTLERGTCVVLHLKEDATEFASVDRLRELATRYSQFMSYPIY 60
+WES ++ ++++ DPRG+TL RGT + L LKE+AT++ ++ ++ L +YSQFM++PIY
Sbjct 222 IWESDSN-EFFVTDDPRGDTLGRGTTITLVLKEEATDYLELETIKNLVRKYSQFMNFPIY 280
Query 61 LKVKQTVSEEVPV 73
+ +T + E P+
Sbjct 281 VWSSKTETVEEPL 293
> mmu:22027 Hsp90b1, ERp99, GRP94, TA-3, Targ2, Tra-1, Tra1, endoplasmin,
gp96; heat shock protein 90, beta (Grp94), member
1; K09487 heat shock protein 90kDa beta
Length=802
Score = 69.3 bits (168), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 30/73 (41%), Positives = 52/73 (71%), Gaps = 1/73 (1%)
Query 1 VWESSADAKYYIDKDPRGNTLERGTCVVLHLKEDATEFASVDRLRELATRYSQFMSYPIY 60
+WES ++ ++ + DPRGNTL RGT + L LKE+A+++ +D ++ L +YSQF+++PIY
Sbjct 222 IWESDSN-EFSVIADPRGNTLGRGTTITLVLKEEASDYLELDTIKNLVRKYSQFINFPIY 280
Query 61 LKVKQTVSEEVPV 73
+ +T + E P+
Sbjct 281 VWSSKTETVEEPL 293
> hsa:7184 HSP90B1, ECGP, GP96, GRP94, TRA1; heat shock protein
90kDa beta (Grp94), member 1; K09487 heat shock protein 90kDa
beta
Length=803
Score = 68.2 bits (165), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 27/61 (44%), Positives = 46/61 (75%), Gaps = 1/61 (1%)
Query 1 VWESSADAKYYIDKDPRGNTLERGTCVVLHLKEDATEFASVDRLRELATRYSQFMSYPIY 60
+WES ++ ++ + DPRGNTL RGT + L LKE+A+++ +D ++ L +YSQF+++PIY
Sbjct 222 IWESDSN-EFSVIADPRGNTLGRGTTITLVLKEEASDYLELDTIKNLVKKYSQFINFPIY 280
Query 61 L 61
+
Sbjct 281 V 281
> bbo:BBOV_III004230 17.m07381; hsp90 protein; K04079 molecular
chaperone HtpG
Length=712
Score = 67.4 bits (163), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 36/92 (39%), Positives = 60/92 (65%), Gaps = 0/92 (0%)
Query 1 VWESSADAKYYIDKDPRGNTLERGTCVVLHLKEDATEFASVDRLRELATRYSQFMSYPIY 60
VWES+A + + KD + L+RGT ++LHLK+D +E+ RL+EL ++S+F+S+PI
Sbjct 150 VWESNASGHFTVTKDESEDQLKRGTRLILHLKDDQSEYLEERRLKELVKKHSEFISFPIR 209
Query 61 LKVKQTVSEEVPVEDEKEDKAEDKEKSEEEDV 92
L V++T EV ++ + +AE K + + DV
Sbjct 210 LSVEKTTETEVTDDEAEPTEAESKPEEKITDV 241
> xla:399408 hsp90b1, ecgp, gp96, grp94, tra1; heat shock protein
90kDa beta (Grp94), member 1
Length=804
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 28/73 (38%), Positives = 53/73 (72%), Gaps = 1/73 (1%)
Query 1 VWESSADAKYYIDKDPRGNTLERGTCVVLHLKEDATEFASVDRLRELATRYSQFMSYPIY 60
+WES ++ ++++ DPRG+TL RG+ + L LKE+AT++ ++ ++ L +YSQF+++PIY
Sbjct 222 IWESDSN-EFFVTDDPRGDTLGRGSTITLVLKEEATDYLELETVKNLVRKYSQFINFPIY 280
Query 61 LKVKQTVSEEVPV 73
+ +T + E P+
Sbjct 281 VWSSKTETVEEPL 293
> cpv:cgd3_3770 Hsp90 ; K04079 molecular chaperone HtpG
Length=711
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 30/71 (42%), Positives = 47/71 (66%), Gaps = 0/71 (0%)
Query 1 VWESSADAKYYIDKDPRGNTLERGTCVVLHLKEDATEFASVDRLRELATRYSQFMSYPIY 60
+WESSA + I D N L+RGT ++LHLKED ++ LR+L ++S+F+S+PI
Sbjct 158 IWESSAGGSFTITNDTSDNKLQRGTRIILHLKEDQLDYLEERTLRDLVKKHSEFISFPIE 217
Query 61 LKVKQTVSEEV 71
L V++T +E+
Sbjct 218 LSVEKTTEKEI 228
> ath:AT5G56030 HSP81-2 (HEAT SHOCK PROTEIN 81-2); ATP binding;
K04079 molecular chaperone HtpG
Length=699
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 30/67 (44%), Positives = 46/67 (68%), Gaps = 0/67 (0%)
Query 1 VWESSADAKYYIDKDPRGNTLERGTCVVLHLKEDATEFASVDRLRELATRYSQFMSYPIY 60
VWES A + + +D G TL RGT +VL+LKED E+ RL++L ++S+F+SYPI
Sbjct 148 VWESQAGGSFTVTRDTSGETLGRGTKMVLYLKEDQLEYLEERRLKDLVKKHSEFISYPIS 207
Query 61 LKVKQTV 67
L +++T+
Sbjct 208 LWIEKTI 214
> dre:386590 hsp90b1, GP96, GRP94, fb61d09, tra-1, tra1, wu:fb61d09,
wu:fq25g01; heat shock protein 90, beta (grp94), member
1; K09487 heat shock protein 90kDa beta
Length=793
Score = 65.1 bits (157), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 27/73 (36%), Positives = 53/73 (72%), Gaps = 1/73 (1%)
Query 1 VWESSADAKYYIDKDPRGNTLERGTCVVLHLKEDATEFASVDRLRELATRYSQFMSYPIY 60
+WES ++ ++ + +DPRG+TL RGT + L +KE+A+++ ++ ++ L +YSQF+++PIY
Sbjct 222 MWESDSN-QFSVIEDPRGDTLGRGTTITLVMKEEASDYLELETIKNLVKKYSQFINFPIY 280
Query 61 LKVKQTVSEEVPV 73
+ +T + E P+
Sbjct 281 VWSSKTETVEEPI 293
> tgo:TGME49_088380 heat shock protein 90 ; K04079 molecular chaperone
HtpG
Length=708
Score = 65.1 bits (157), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 33/84 (39%), Positives = 57/84 (67%), Gaps = 1/84 (1%)
Query 1 VWESSADAKYYIDK-DPRGNTLERGTCVVLHLKEDATEFASVDRLRELATRYSQFMSYPI 59
VWESSA + + K + + + RGT ++LH+KED TE+ RL++L ++S+F+S+PI
Sbjct 148 VWESSAGGSFTVSKAEGQFENIVRGTRIILHMKEDQTEYLEDRRLKDLVKKHSEFISFPI 207
Query 60 YLKVKQTVSEEVPVEDEKEDKAED 83
L V+++V +E+ +++E AED
Sbjct 208 ELAVEKSVDKEITESEDEEKPAED 231
> ath:AT5G56010 HSP81-3; ATP binding / unfolded protein binding;
K04079 molecular chaperone HtpG
Length=699
Score = 64.7 bits (156), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 29/67 (43%), Positives = 45/67 (67%), Gaps = 0/67 (0%)
Query 1 VWESSADAKYYIDKDPRGNTLERGTCVVLHLKEDATEFASVDRLRELATRYSQFMSYPIY 60
VWES A + + +D G L RGT +VL+LKED E+ RL++L ++S+F+SYPI
Sbjct 148 VWESQAGGSFTVTRDTSGEALGRGTKMVLYLKEDQMEYIEERRLKDLVKKHSEFISYPIS 207
Query 61 LKVKQTV 67
L +++T+
Sbjct 208 LWIEKTI 214
> ath:AT5G56000 heat shock protein 81-4 (HSP81-4); K04079 molecular
chaperone HtpG
Length=699
Score = 64.7 bits (156), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 28/67 (41%), Positives = 45/67 (67%), Gaps = 0/67 (0%)
Query 1 VWESSADAKYYIDKDPRGNTLERGTCVVLHLKEDATEFASVDRLRELATRYSQFMSYPIY 60
VWES A + + +D G L RGT ++L+LKED E+ RL++L ++S+F+SYPI
Sbjct 148 VWESQAGGSFTVTRDTSGEALGRGTKMILYLKEDQMEYIEERRLKDLVKKHSEFISYPIS 207
Query 61 LKVKQTV 67
L +++T+
Sbjct 208 LWIEKTI 214
> ath:AT3G07770 ATP binding
Length=799
Score = 62.4 bits (150), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 33/86 (38%), Positives = 55/86 (63%), Gaps = 4/86 (4%)
Query 1 VWESSADAKYYI---DKDPRGNTLERGTCVVLHLKEDATEFASVDRLRELATRYSQFMSY 57
VWE A++ + D DP+ + + RGT + LHLK++A FA +R+++L YSQF+S+
Sbjct 243 VWEGEANSSSFTIQEDTDPQ-SLIPRGTRITLHLKQEAKNFADPERIQKLVKNYSQFVSF 301
Query 58 PIYLKVKQTVSEEVPVEDEKEDKAED 83
PIY ++ ++EV VED+ + +D
Sbjct 302 PIYTWQEKGYTKEVEVEDDPTETKKD 327
> dre:30591 hsp90a.1, fb17b01, hsp90, hsp90a, hsp90alpha, wu:fb17b01,
zgc:86652; heat shock protein 90-alpha 1; K04079 molecular
chaperone HtpG
Length=726
Score = 62.0 bits (149), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 35/98 (35%), Positives = 62/98 (63%), Gaps = 7/98 (7%)
Query 1 VWESSADAKYYIDKDPRGNTLERGTCVVLHLKEDATEFASVDRLRELATRYSQFMSYPIY 60
+WES+A + + D G ++ RGT V+LHLKED +E+ R++E+ ++SQF+ YPI
Sbjct 159 IWESAAGGSFTVKPD-FGESIGRGTKVILHLKEDQSEYVEEKRIKEVVKKHSQFIGYPIT 217
Query 61 LKVKQTVSEEVPVEDEKEDK------AEDKEKSEEEDV 92
L +++ +EV +E+ ++ + EDK+K + ED+
Sbjct 218 LYIEKQREKEVDLEEGEKQEEEEVAAGEDKDKPKIEDL 255
> mmu:15519 Hsp90aa1, 86kDa, 89kDa, AL024080, AL024147, Hsp86-1,
Hsp89, Hsp90, Hspca, hsp4; heat shock protein 90, alpha (cytosolic),
class A member 1; K04079 molecular chaperone HtpG
Length=733
Score = 61.6 bits (148), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 29/62 (46%), Positives = 40/62 (64%), Gaps = 1/62 (1%)
Query 2 WESSADAKYYIDKDPRGNTLERGTCVVLHLKEDATEFASVDRLRELATRYSQFMSYPIYL 61
WESSA + + D G + RGT V+LHLKED TE+ R++E+ ++SQF+ YPI L
Sbjct 162 WESSAGGSFTVRTD-TGEPMGRGTKVILHLKEDQTEYLEERRIKEIVKKHSQFIGYPITL 220
Query 62 KV 63
V
Sbjct 221 FV 222
> hsa:3320 HSP90AA1, FLJ31884, HSP86, HSP89A, HSP90A, HSP90N,
HSPC1, HSPCA, HSPCAL1, HSPCAL4, HSPN, Hsp89, Hsp90, LAP2; heat
shock protein 90kDa alpha (cytosolic), class A member 1;
K04079 molecular chaperone HtpG
Length=854
Score = 60.8 bits (146), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 29/62 (46%), Positives = 40/62 (64%), Gaps = 1/62 (1%)
Query 2 WESSADAKYYIDKDPRGNTLERGTCVVLHLKEDATEFASVDRLRELATRYSQFMSYPIYL 61
WESSA + + D G + RGT V+LHLKED TE+ R++E+ ++SQF+ YPI L
Sbjct 284 WESSAGGSFTVRTD-TGEPMGRGTKVILHLKEDQTEYLEERRIKEIVKKHSQFIGYPITL 342
Query 62 KV 63
V
Sbjct 343 FV 344
> ath:AT5G52640 ATHSP90.1 (HEAT SHOCK PROTEIN 90.1); ATP binding
/ unfolded protein binding; K04079 molecular chaperone HtpG
Length=705
Score = 60.8 bits (146), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 28/66 (42%), Positives = 42/66 (63%), Gaps = 0/66 (0%)
Query 1 VWESSADAKYYIDKDPRGNTLERGTCVVLHLKEDATEFASVDRLRELATRYSQFMSYPIY 60
VWES A + + +D G L RGT + L LK+D E+ RL++L ++S+F+SYPIY
Sbjct 153 VWESQAGGSFTVTRDVDGEPLGRGTKITLFLKDDQLEYLEERRLKDLVKKHSEFISYPIY 212
Query 61 LKVKQT 66
L ++T
Sbjct 213 LWTEKT 218
> dre:30573 hsp90ab1, hsp90b, hsp90beta, wu:fa29f01, wu:fa91e11,
wu:fd59e11, wu:gcd22h07; heat shock protein 90, alpha (cytosolic),
class B member 1; K04079 molecular chaperone HtpG
Length=725
Score = 60.8 bits (146), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 29/62 (46%), Positives = 40/62 (64%), Gaps = 1/62 (1%)
Query 2 WESSADAKYYIDKDPRGNTLERGTCVVLHLKEDATEFASVDRLRELATRYSQFMSYPIYL 61
WESSA + + D G + RGT V+LHLKED TE+ R++E+ ++SQF+ YPI L
Sbjct 156 WESSAGGSFTVKVD-HGEPIGRGTKVILHLKEDQTEYIEEKRVKEVVKKHSQFIGYPITL 214
Query 62 KV 63
V
Sbjct 215 YV 216
> pfa:PF07_0029 heat shock protein 86; K04079 molecular chaperone
HtpG
Length=745
Score = 60.5 bits (145), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 26/61 (42%), Positives = 40/61 (65%), Gaps = 0/61 (0%)
Query 1 VWESSADAKYYIDKDPRGNTLERGTCVVLHLKEDATEFASVDRLRELATRYSQFMSYPIY 60
VWES+A + + KD L RGT ++LHLKED E+ R+++L ++S+F+S+PI
Sbjct 147 VWESAAGGSFTVTKDETNEKLGRGTKIILHLKEDQLEYLEEKRIKDLVKKHSEFISFPIK 206
Query 61 L 61
L
Sbjct 207 L 207
> mmu:15516 Hsp90ab1, 90kDa, AL022974, C81438, Hsp84, Hsp84-1,
Hsp90, Hspcb, MGC115780; heat shock protein 90 alpha (cytosolic),
class B member 1; K04079 molecular chaperone HtpG
Length=724
Score = 59.7 bits (143), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 28/60 (46%), Positives = 39/60 (65%), Gaps = 1/60 (1%)
Query 2 WESSADAKYYIDKDPRGNTLERGTCVVLHLKEDATEFASVDRLRELATRYSQFMSYPIYL 61
WESSA + + D G + RGT V+LHLKED TE+ R++E+ ++SQF+ YPI L
Sbjct 157 WESSAGGSFTVRAD-HGEPIGRGTKVILHLKEDQTEYLEERRVKEVVKKHSQFIGYPITL 215
> hsa:3326 HSP90AB1, D6S182, FLJ26984, HSP90-BETA, HSP90B, HSPC2,
HSPCB; heat shock protein 90kDa alpha (cytosolic), class
B member 1; K04079 molecular chaperone HtpG
Length=724
Score = 59.7 bits (143), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 28/60 (46%), Positives = 39/60 (65%), Gaps = 1/60 (1%)
Query 2 WESSADAKYYIDKDPRGNTLERGTCVVLHLKEDATEFASVDRLRELATRYSQFMSYPIYL 61
WESSA + + D G + RGT V+LHLKED TE+ R++E+ ++SQF+ YPI L
Sbjct 157 WESSAGGSFTVRAD-HGEPIGRGTKVILHLKEDQTEYLEERRVKEVVKKHSQFIGYPITL 215
> ath:AT4G24190 SHD; SHD (SHEPHERD); ATP binding / unfolded protein
binding
Length=823
Score = 59.7 bits (143), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 26/61 (42%), Positives = 41/61 (67%), Gaps = 0/61 (0%)
Query 1 VWESSADAKYYIDKDPRGNTLERGTCVVLHLKEDATEFASVDRLRELATRYSQFMSYPIY 60
VWES A+ K+ + +D L RGT + LHL+++A E+ +L+EL RYS+F+++PI
Sbjct 222 VWESKANGKFAVSEDTWNEPLGRGTEIRLHLRDEAGEYLEESKLKELVKRYSEFINFPIS 281
Query 61 L 61
L
Sbjct 282 L 282
> sce:YMR186W HSC82, HSP90; Cytoplasmic chaperone of the Hsp90
family, redundant in function and nearly identical with Hsp82p,
and together they are essential; expressed constitutively
at 10-fold higher basal levels than HSP82 and induced 2-3
fold by heat shock; K04079 molecular chaperone HtpG
Length=705
Score = 58.5 bits (140), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 27/73 (36%), Positives = 45/73 (61%), Gaps = 0/73 (0%)
Query 1 VWESSADAKYYIDKDPRGNTLERGTCVVLHLKEDATEFASVDRLRELATRYSQFMSYPIY 60
+WES+A + + D + RGT + L LK+D E+ R++E+ R+S+F++YPI
Sbjct 147 IWESNAGGSFTVTLDEVNERIGRGTVLRLFLKDDQLEYLEEKRIKEVIKRHSEFVAYPIQ 206
Query 61 LKVKQTVSEEVPV 73
L V + V +EVP+
Sbjct 207 LLVTKEVEKEVPI 219
> xla:446459 hsp90ab1, hsp90b, hsp90beta; heat shock protein 90kDa
alpha (cytosolic), class B member 1; K04079 molecular chaperone
HtpG
Length=722
Score = 58.5 bits (140), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 28/60 (46%), Positives = 38/60 (63%), Gaps = 1/60 (1%)
Query 2 WESSADAKYYIDKDPRGNTLERGTCVVLHLKEDATEFASVDRLRELATRYSQFMSYPIYL 61
WESSA + + D G + RGT V+LHLKED TE+ R++E ++SQF+ YPI L
Sbjct 157 WESSAGGSFTVKVD-TGEPIGRGTKVILHLKEDQTEYLEEKRVKETVKKHSQFIGYPITL 215
> sce:YPL240C HSP82, HSP90; Hsp82p; K04079 molecular chaperone
HtpG
Length=709
Score = 58.5 bits (140), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 27/73 (36%), Positives = 45/73 (61%), Gaps = 0/73 (0%)
Query 1 VWESSADAKYYIDKDPRGNTLERGTCVVLHLKEDATEFASVDRLRELATRYSQFMSYPIY 60
+WES+A + + D + RGT + L LK+D E+ R++E+ R+S+F++YPI
Sbjct 147 IWESNAGGSFTVTLDEVNERIGRGTILRLFLKDDQLEYLEEKRIKEVIKRHSEFVAYPIQ 206
Query 61 LKVKQTVSEEVPV 73
L V + V +EVP+
Sbjct 207 LVVTKEVEKEVPI 219
> dre:565155 hsp90a.2, cb820, hsp90a2; heat shock protein 90-alpha
2; K04079 molecular chaperone HtpG
Length=734
Score = 58.2 bits (139), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 30/70 (42%), Positives = 44/70 (62%), Gaps = 1/70 (1%)
Query 2 WESSADAKYYIDKDPRGNTLERGTCVVLHLKEDATEFASVDRLRELATRYSQFMSYPIYL 61
WESSA + + D + RGT V+LHLKED TE+ R++E+ ++SQF+ YPI L
Sbjct 160 WESSAGGSFTVKVD-NSEPIGRGTKVILHLKEDQTEYIEERRIKEIVKKHSQFIGYPITL 218
Query 62 KVKQTVSEEV 71
V++ +EV
Sbjct 219 FVEKERDKEV 228
> xla:444024 hsp90aa1.1, MGC82579, hsp86, hsp89, hsp90, hsp90a,
hspc1, hspca, hspn, lap2; heat shock protein 90kDa alpha (cytosolic),
class A member 1, gene 1; K04079 molecular chaperone
HtpG
Length=729
Score = 57.8 bits (138), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 28/62 (45%), Positives = 39/62 (62%), Gaps = 1/62 (1%)
Query 2 WESSADAKYYIDKDPRGNTLERGTCVVLHLKEDATEFASVDRLRELATRYSQFMSYPIYL 61
WESSA + + D L RGT V+LHLKED +E+ R++E+ ++SQF+ YPI L
Sbjct 163 WESSAGGSFTVRVD-NSEPLGRGTKVILHLKEDQSEYFEEKRIKEIVKKHSQFIGYPITL 221
Query 62 KV 63
V
Sbjct 222 FV 223
> ath:AT2G04030 CR88; CR88; ATP binding
Length=780
Score = 56.2 bits (134), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 30/64 (46%), Positives = 40/64 (62%), Gaps = 5/64 (7%)
Query 1 VWESSADAKYYI---DKDPRGNTLERGTCVVLHLKED-ATEFASVDRLRELATRYSQFMS 56
VWES AD+ Y+ + DP N L RGT + L+L+ED EFA R++ L YSQF+
Sbjct 225 VWESVADSSSYLIREETDPD-NILRRGTQITLYLREDDKYEFAESTRIKNLVKNYSQFVG 283
Query 57 YPIY 60
+PIY
Sbjct 284 FPIY 287
> tpv:TP01_0934 heat shock protein 90
Length=1009
Score = 54.7 bits (130), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 26/61 (42%), Positives = 36/61 (59%), Gaps = 0/61 (0%)
Query 1 VWESSADAKYYIDKDPRGNTLERGTCVVLHLKEDATEFASVDRLRELATRYSQFMSYPIY 60
VW SSA Y + +D + + GT + L L+EDAT++ D L L +YSQF+ YPI
Sbjct 224 VWRSSAANSYELYEDTDNSLGDHGTLITLELREDATDYLKTDVLENLVKKYSQFVKYPIQ 283
Query 61 L 61
L
Sbjct 284 L 284
> dre:571959 trap1, fc85a11, wu:fc85a11; TNF receptor-associated
protein 1; K09488 TNF receptor-associated protein 1
Length=719
Score = 54.7 bits (130), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 24/60 (40%), Positives = 40/60 (66%), Gaps = 3/60 (5%)
Query 2 WESSADAKYYIDKDPRGNTLERGTCVVLHLKEDATEFASVDRLRELATRYSQFMSYPIYL 61
W S + + + + + +GT +VLHLK+D EF+S DR++E+ T+YS F+S+PI+L
Sbjct 246 WSSDGSGVFEVAE---ASGVRQGTKIVLHLKDDCKEFSSEDRVKEVVTKYSNFVSFPIFL 302
> tgo:TGME49_092920 heat shock protein 90, putative ; K09488 TNF
receptor-associated protein 1
Length=861
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 20/42 (47%), Positives = 34/42 (80%), Gaps = 0/42 (0%)
Query 21 LERGTCVVLHLKEDATEFASVDRLRELATRYSQFMSYPIYLK 62
L+RGT +V HLK+D EF+++ ++E AT++S F+++PIY+K
Sbjct 331 LKRGTKIVCHLKKDCLEFSNIHHVKECATKFSSFVNFPIYVK 372
> cel:C47E8.5 daf-21; abnormal DAuer Formation family member (daf-21);
K04079 molecular chaperone HtpG
Length=702
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 35/93 (37%), Positives = 57/93 (61%), Gaps = 5/93 (5%)
Query 2 WESSADAKYYID--KDPRGNTLERGTCVVLHLKEDATEFASVDRLRELATRYSQFMSYPI 59
WESSA + + DP + RGT +V+H+KED +F +++E+ ++SQF+ YPI
Sbjct 150 WESSAGGSFVVRPFNDPE---VTRGTKIVMHIKEDQIDFLEERKIKEIVKKHSQFIGYPI 206
Query 60 YLKVKQTVSEEVPVEDEKEDKAEDKEKSEEEDV 92
L V++ +EV E+ E K E+K++ E E+V
Sbjct 207 KLVVEKEREKEVEDEEAVEAKDEEKKEGEVENV 239
> hsa:10131 TRAP1, HSP75, HSP90L; TNF receptor-associated protein
1; K09488 TNF receptor-associated protein 1
Length=704
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 19/38 (50%), Positives = 30/38 (78%), Gaps = 0/38 (0%)
Query 24 GTCVVLHLKEDATEFASVDRLRELATRYSQFMSYPIYL 61
GT +++HLK D EF+S R+R++ T+YS F+S+P+YL
Sbjct 250 GTKIIIHLKSDCKEFSSEARVRDVVTKYSNFVSFPLYL 287
> mmu:68015 Trap1, 2410002K23Rik, HSP75; TNF receptor-associated
protein 1; K09488 TNF receptor-associated protein 1
Length=706
Score = 49.7 bits (117), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 19/42 (45%), Positives = 32/42 (76%), Gaps = 0/42 (0%)
Query 24 GTCVVLHLKEDATEFASVDRLRELATRYSQFMSYPIYLKVKQ 65
GT +++HLK D +FAS R++++ T+YS F+S+P+YL K+
Sbjct 252 GTKIIIHLKSDCKDFASESRVQDVVTKYSNFVSFPLYLNGKR 293
> pfa:PF14_0417 HSP90
Length=927
Score = 46.2 bits (108), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 31/104 (29%), Positives = 53/104 (50%), Gaps = 25/104 (24%)
Query 24 GTCVVLHLKEDATEFASVDRLRELATRYSQFMSYPIYLKVKQTVSEEVPVEDEKEDKAED 83
GT ++LHLKE+ E+ +L+EL +YS+F+ +PI + SE++ D
Sbjct 307 GTKIILHLKEECDEYLEDYKLKELIKKYSEFIKFPI-----EIWSEKI-----------D 350
Query 84 KEKSEEEDVQVQDVDEETDDKEAKKKKTKTVEREVERWDRINEQ 127
E+ ++ V ++D D K K KT+ + W++IN Q
Sbjct 351 YERVPDDSVSLKDGD---------KMKMKTITKRYHEWEKINVQ 385
> bbo:BBOV_IV010880 23.m05763; heat shock protein 75; K09488 TNF
receptor-associated protein 1
Length=623
Score = 44.7 bits (104), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 15/42 (35%), Positives = 31/42 (73%), Gaps = 0/42 (0%)
Query 21 LERGTCVVLHLKEDATEFASVDRLRELATRYSQFMSYPIYLK 62
L RGT +V HL++D FA+ ++++A ++S F+++P+Y++
Sbjct 187 LPRGTKIVCHLRDDCVVFANTANVKKVAEKFSAFVNFPLYIQ 228
> pfa:PF11_0188 heat shock protein 90, putative
Length=930
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 19/74 (25%), Positives = 43/74 (58%), Gaps = 2/74 (2%)
Query 20 TLERGTCVVLHLKEDATEFASVDRLRELATRYSQFMSYPIY-LKVKQTVSEEVPVE-DEK 77
+ +GT ++ HLK+ EF+++ ++++ ++S F+++P+Y LK K+ + +E +E
Sbjct 268 NIPKGTKIICHLKDSCKEFSNIQNVQKIVEKFSSFINFPVYVLKKKKILQTRKDIEQNEL 327
Query 78 EDKAEDKEKSEEED 91
D + E + D
Sbjct 328 NDHTQQNELNHHND 341
> tpv:TP01_0753 heat shock protein 75; K09488 TNF receptor-associated
protein 1
Length=724
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 19/69 (27%), Positives = 44/69 (63%), Gaps = 3/69 (4%)
Query 21 LERGTCVVLHLKEDATEFASVDRLRELATRYSQFMSYPIYLKVKQT---VSEEVPVEDEK 77
L RGT ++ +LK+D+ F + + ++++A ++S F+++P++L+ K ++ + P+ EK
Sbjct 248 LPRGTKIICYLKDDSLLFCNSNNVKKVAEKFSSFINFPLFLQEKDKDVEITTQKPLWVEK 307
Query 78 EDKAEDKEK 86
+ E+ K
Sbjct 308 KSSPEEHTK 316
> tpv:TP04_0646 heat shock protein 90
Length=913
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 23/75 (30%), Positives = 44/75 (58%), Gaps = 4/75 (5%)
Query 2 WESSADAKYYI----DKDPRGNTLERGTCVVLHLKEDATEFASVDRLRELATRYSQFMSY 57
W+S ++ Y I +++ ++ GT +VLHLK + ++ +L+EL +YS+F+ +
Sbjct 277 WKSDSNGTYTIGRVENQELNDKFMKSGTRIVLHLKPECDDYLEDYKLKELLRKYSEFIRF 336
Query 58 PIYLKVKQTVSEEVP 72
PI + V++ E VP
Sbjct 337 PIQVWVERIEYERVP 351
> tgo:TGME49_110430 heat shock protein 90, putative (EC:3.2.1.3
2.7.13.3)
Length=1100
Score = 41.6 bits (96), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 34/128 (26%), Positives = 59/128 (46%), Gaps = 24/128 (18%)
Query 24 GTCVVLHLKEDATEFASVDRLRELATRYSQFMSYPIYLKVKQTVSEEVPVEDEK------ 77
GT VVLHLKED+ ++ +L+EL +YS+F+ PI++ ++ E VP E+
Sbjct 484 GTRVVLHLKEDSDDYLEDYKLKELMRKYSEFIQLPIHIWSERIEYERVPEGSEQAAAPPA 543
Query 78 EDKAEDKEKSEEEDVQVQDVDEE----TDDKEAKKK--------------KTKTVEREVE 119
+ D + + +++DE+ D A+++ K KTV
Sbjct 544 SSASADGQMPGLTRLTPENIDEQLGKVAGDPGARRQSEPGFPSSTGAAGTKFKTVTHRYY 603
Query 120 RWDRINEQ 127
W+ +N Q
Sbjct 604 EWEHLNTQ 611
> bbo:BBOV_III007380 17.m07646; heat shock protein 90
Length=795
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 28/130 (21%), Positives = 59/130 (45%), Gaps = 30/130 (23%)
Query 2 WESSADAKYYI----DKDPRGNTLERGTCVVLHLKEDATEFASVDRLRELATRYSQFMSY 57
W+S + + + D + + ++ GT +VLH+K + ++ +++EL +YS+F+ +
Sbjct 268 WKSETNGTFSVAQVNDDELQKGFMKCGTRIVLHIKPECDDYLEDYKIKELLRKYSEFVRF 327
Query 58 PIYLKVKQTVSEEVPVEDEKEDKAEDKEKSEEEDVQVQDVDEETDDKEAKKKKTKTVERE 117
PI + V++ E VP +E+ E K + KT+ ++
Sbjct 328 PIQVWVEKVEYERVP--------------------------DESTAVEGKPGRYKTISKK 361
Query 118 VERWDRINEQ 127
W+ +N Q
Sbjct 362 RHEWEHVNTQ 371
> eco:b0473 htpG, ECK0467, JW0462; molecular chaperone HSP90 family;
K04079 molecular chaperone HtpG
Length=624
Score = 37.4 bits (85), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 23/82 (28%), Positives = 45/82 (54%), Gaps = 3/82 (3%)
Query 1 VWESSADAKYYIDKDPRGNTLERGTCVVLHLKEDATEFASVDRLRELATRYSQFMSYPIY 60
WES+ + +Y + + + RGT + LHL+E EF R+R + ++YS ++ P+
Sbjct 153 FWESAGEGEYTVADITKED---RGTEITLHLREGEDEFLDDWRVRSIISKYSDHIALPVE 209
Query 61 LKVKQTVSEEVPVEDEKEDKAE 82
++ ++ E + EK +KA+
Sbjct 210 IEKREEKDGETVISWEKINKAQ 231
> hsa:10291 SF3A1, PRP21, PRPF21, SAP114, SF3A120; splicing factor
3a, subunit 1, 120kDa; K12825 splicing factor 3A subunit
1
Length=728
Score = 30.4 bits (67), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 15/33 (45%), Positives = 24/33 (72%), Gaps = 0/33 (0%)
Query 77 KEDKAEDKEKSEEEDVQVQDVDEETDDKEAKKK 109
K++KAE+ ++D QVQD+DE +DD+E +K
Sbjct 270 KQEKAEEPPSQLDQDTQVQDMDEGSDDEEEGQK 302
> mmu:67465 Sf3a1, 1200014H24Rik, 5930416L09Rik, AI159724; splicing
factor 3a, subunit 1; K12825 splicing factor 3A subunit
1
Length=791
Score = 28.5 bits (62), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 14/32 (43%), Positives = 23/32 (71%), Gaps = 0/32 (0%)
Query 78 EDKAEDKEKSEEEDVQVQDVDEETDDKEAKKK 109
++KAE+ ++D QVQD+DE +DD+E +K
Sbjct 334 QEKAEETPSQLDQDTQVQDMDEGSDDEEEGQK 365
> xla:444135 MGC80562 protein; K12825 splicing factor 3A subunit
1
Length=802
Score = 27.7 bits (60), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 14/33 (42%), Positives = 22/33 (66%), Gaps = 0/33 (0%)
Query 77 KEDKAEDKEKSEEEDVQVQDVDEETDDKEAKKK 109
K+ K+E+ ++D QVQD+DE +DD+E K
Sbjct 329 KDKKSEESSSQMDQDTQVQDMDEGSDDEEYTSK 361
Lambda K H
0.305 0.125 0.339
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2054672932
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40