bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2445_orf1
Length=110
Score E
Sequences producing significant alignments: (Bits) Value
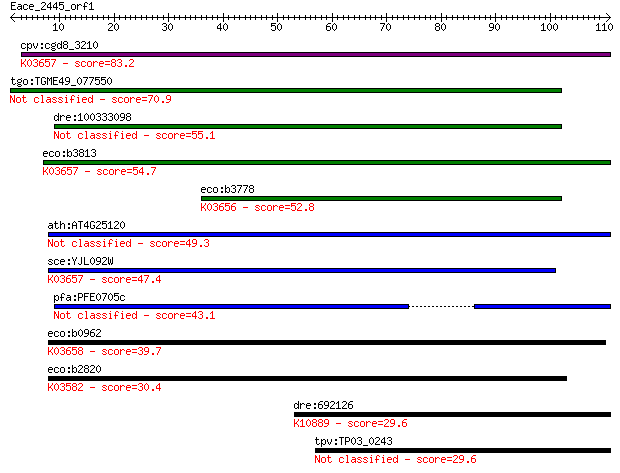
cpv:cgd8_3210 UvrD like super family I helicase involved in nu... 83.2 2e-16
tgo:TGME49_077550 uvrD/REP helicase domain-containing protein ... 70.9 1e-12
dre:100333098 hypothetical protein LOC100333098 55.1 6e-08
eco:b3813 uvrD, dar-2, dda, ECK3808, JW3786, mutU, pdeB, rad, ... 54.7 8e-08
eco:b3778 rep, dasC, ECK3770, JW5604, mbrA, mmrA?; DNA helicas... 52.8 3e-07
ath:AT4G25120 ATP binding / ATP-dependent DNA helicase/ DNA bi... 49.3 3e-06
sce:YJL092W SRS2, HPR5; DNA helicase and DNA-dependent ATPase ... 47.4 1e-05
pfa:PFE0705c helicase, belonging to UvrD family, putative 43.1 2e-04
eco:b0962 helD, ECK0953, JW0945, srjB; DNA helicase IV (EC:3.6... 39.7 0.002
eco:b2820 recB, ECK2816, ior, JW2788, rorA; exonuclease V (Rec... 30.4 1.5
dre:692126 fancb; Fanconi anemia, complementation group B; K10... 29.6 2.2
tpv:TP03_0243 hypothetical protein 29.6 2.6
> cpv:cgd8_3210 UvrD like super family I helicase involved in
nucleotide excision repair, possible bacterial horizontal transfer
; K03657 DNA helicase II / ATP-dependent DNA helicase
PcrA [EC:3.6.4.12]
Length=917
Score = 83.2 bits (204), Expect = 2e-16, Method: Composition-based stats.
Identities = 46/109 (42%), Positives = 59/109 (54%), Gaps = 4/109 (3%)
Query 3 QQPPLLDFTDLTVKALKLLEPPAT-RAELGAQWPYLVVDEFQDTNSNQFKFICLLALQRP 61
+P L+D+TDL AL LLE R ++ +PY+ DEFQDT+ QFK + LL
Sbjct 296 HKPYLVDYTDLITLALNLLENNINIREKIQDSYPYIFCDEFQDTSKLQFKILELLT---K 352
Query 62 PNIQTLHTAVGNPTLPNSTPGRGGVTVVGDDDQSIYKWRGTHTGIFHEF 110
I + RGG+TVVGDDDQ+IY WRG TG+F F
Sbjct 353 SIINKKRKFEAEKFTRDQNLRRGGITVVGDDDQAIYSWRGVETGVFSRF 401
> tgo:TGME49_077550 uvrD/REP helicase domain-containing protein
(EC:3.1.11.5 3.4.21.72)
Length=2851
Score = 70.9 bits (172), Expect = 1e-12, Method: Composition-based stats.
Identities = 54/123 (43%), Positives = 67/123 (54%), Gaps = 22/123 (17%)
Query 1 REQQPPLLDFTDLTVKALKLLEP-PATRAELGAQWPYLVVDEFQDTNSNQFKFICLLALQ 59
R+Q PPLLD +DL + L LLE P R +L +P +VVDEFQDT+ QFK + LAL
Sbjct 1178 RQQTPPLLDCSDLILLTLALLESQPNIRRDLATAYPIVVVDEFQDTSLPQFKVVKALALG 1237
Query 60 RPPNIQT-------LHTAV-------------GNPTLPNST-PGRGGVTVVGDDDQSIYK 98
R + +H + GNP +T RGGVTVVGDDDQSIY
Sbjct 1238 RQEQEREQARERAWMHETMERERGEGRAGFCAGNPGRQEATRQRRGGVTVVGDDDQSIYG 1297
Query 99 WRG 101
+RG
Sbjct 1298 FRG 1300
> dre:100333098 hypothetical protein LOC100333098
Length=827
Score = 55.1 bits (131), Expect = 6e-08, Method: Composition-based stats.
Identities = 34/94 (36%), Positives = 48/94 (51%), Gaps = 20/94 (21%)
Query 9 DFTDLTVKALKLLE-PPATRAELGAQWPYLVVDEFQDTNSNQFKFICLLALQRPPNIQTL 67
DF DL + + L P + ++ Y++VDE+QDTN+ Q+ ++ LLA QRP ++
Sbjct 240 DFGDLLLHPIDLFRRNPDVLKDYHQRFRYILVDEYQDTNTAQYMWLRLLA-QRPKDVPQ- 297
Query 68 HTAVGNPTLPNSTPGRGGVTVVGDDDQSIYKWRG 101
V VGDDDQSIY WRG
Sbjct 298 -----------------NVCCVGDDDQSIYGWRG 314
> eco:b3813 uvrD, dar-2, dda, ECK3808, JW3786, mutU, pdeB, rad,
recL, srjC, uvr502, uvrE; DNA-dependent ATPase I and helicase
II (EC:3.6.1.-); K03657 DNA helicase II / ATP-dependent
DNA helicase PcrA [EC:3.6.4.12]
Length=720
Score = 54.7 bits (130), Expect = 8e-08, Method: Composition-based stats.
Identities = 37/105 (35%), Positives = 51/105 (48%), Gaps = 24/105 (22%)
Query 7 LLDFTDLTVKALKL-LEPPATRAELGAQWPYLVVDEFQDTNSNQFKFICLLALQRPPNIQ 65
L+DF +L ++A +L L P ++ ++VDEFQDTN+ Q+ +I LLA
Sbjct 186 LVDFAELLLRAHELWLNKPHILQHYRERFTNILVDEFQDTNNIQYAWIRLLA-------- 237
Query 66 TLHTAVGNPTLPNSTPGRGGVTVVGDDDQSIYKWRGTHTGIFHEF 110
G+ G V +VGDDDQSIY WRG F
Sbjct 238 ------GDT---------GKVMIVGDDDQSIYGWRGAQVENIQRF 267
> eco:b3778 rep, dasC, ECK3770, JW5604, mbrA, mmrA?; DNA helicase
and single-stranded DNA-dependent ATPase (EC:3.6.1.-); K03656
ATP-dependent DNA helicase Rep [EC:3.6.4.12]
Length=673
Score = 52.8 bits (125), Expect = 3e-07, Method: Composition-based stats.
Identities = 29/66 (43%), Positives = 37/66 (56%), Gaps = 23/66 (34%)
Query 36 YLVVDEFQDTNSNQFKFICLLALQRPPNIQTLHTAVGNPTLPNSTPGRGGVTVVGDDDQS 95
YL+VDE+QDTN++Q++ + LL VG+ R TVVGDDDQS
Sbjct 210 YLLVDEYQDTNTSQYELVKLL--------------VGS---------RARFTVVGDDDQS 246
Query 96 IYKWRG 101
IY WRG
Sbjct 247 IYSWRG 252
> ath:AT4G25120 ATP binding / ATP-dependent DNA helicase/ DNA
binding / hydrolase
Length=1149
Score = 49.3 bits (116), Expect = 3e-06, Method: Composition-based stats.
Identities = 31/104 (29%), Positives = 46/104 (44%), Gaps = 25/104 (24%)
Query 8 LDFTDLTVKALKLLEP-PATRAELGAQWPYLVVDEFQDTNSNQFKFICLLALQRPPNIQT 66
LD+ DL ++ LL P E W ++VDEFQDT++ Q+K + +L
Sbjct 450 LDYHDLISCSVTLLSDFPEVFKECQDTWKAIIVDEFQDTSTMQYKLLRMLG--------- 500
Query 67 LHTAVGNPTLPNSTPGRGGVTVVGDDDQSIYKWRGTHTGIFHEF 110
+T+VGDDDQSI+ + G + F F
Sbjct 501 ---------------SHNHITIVGDDDQSIFGFNGADSSGFDSF 529
> sce:YJL092W SRS2, HPR5; DNA helicase and DNA-dependent ATPase
involved in DNA repair, needed for proper timing of commitment
to meiotic recombination and transition from Meiosis I
to II; blocks trinucleotide repeat expansion; affects genome
stability (EC:3.6.1.-); K03657 DNA helicase II / ATP-dependent
DNA helicase PcrA [EC:3.6.4.12]
Length=1174
Score = 47.4 bits (111), Expect = 1e-05, Method: Composition-based stats.
Identities = 34/93 (36%), Positives = 45/93 (48%), Gaps = 23/93 (24%)
Query 8 LDFTDLTVKALKLLEPPATRAELGAQWPYLVVDEFQDTNSNQFKFICLLALQRPPNIQTL 67
LDF DL + +LL TR + + +++VDEFQDTN Q + L A
Sbjct 217 LDFDDLLMYTFRLL----TRVRVLSNIKHVLVDEFQDTNGIQLDLMFLFA---------- 262
Query 68 HTAVGNPTLPNSTPGRGGVTVVGDDDQSIYKWR 100
GN L G+T+VGD DQSIY +R
Sbjct 263 ---KGNHHLSR------GMTIVGDPDQSIYAFR 286
> pfa:PFE0705c helicase, belonging to UvrD family, putative
Length=1441
Score = 43.1 bits (100), Expect = 2e-04, Method: Composition-based stats.
Identities = 23/68 (33%), Positives = 39/68 (57%), Gaps = 4/68 (5%)
Query 9 DFTDLTVKALKLLEPPA-TRAELGAQWPYLVVDEFQDTNSNQFKFICLLALQRPPNI--Q 65
DF DL ++ +L++ A R ++ +W Y+ DEFQDTN+ QF + P+ Q
Sbjct 439 DFDDLLIETYRLMKDNADIRNKILEEWNYVFCDEFQDTNTTQFNILQFFVNHNVPSTLDQ 498
Query 66 TLHTAVGN 73
+++T+ GN
Sbjct 499 SIYTS-GN 505
Score = 42.4 bits (98), Expect = 4e-04, Method: Composition-based stats.
Identities = 15/25 (60%), Positives = 21/25 (84%), Gaps = 0/25 (0%)
Query 86 VTVVGDDDQSIYKWRGTHTGIFHEF 110
+TV+GDDDQSIY +RG H +F++F
Sbjct 685 LTVIGDDDQSIYSFRGAHINVFYKF 709
> eco:b0962 helD, ECK0953, JW0945, srjB; DNA helicase IV (EC:3.6.1.-);
K03658 DNA helicase IV [EC:3.6.4.12]
Length=684
Score = 39.7 bits (91), Expect = 0.002, Method: Composition-based stats.
Identities = 33/105 (31%), Positives = 48/105 (45%), Gaps = 28/105 (26%)
Query 8 LDFTDLTVKALKLLEPPATRAELGAQWPYLVVDEFQDTNSNQFKFICLLALQRPPNIQTL 67
+DF+ L +A+ +LE + + W +++VDEFQD + + LLA R N QT
Sbjct 409 VDFSGLIHQAIVILE----KGRFISPWKHILVDEFQDISPQR---AALLAALRKQNSQTT 461
Query 68 HTAVGNPTLPNSTPGRGGVTVVGDDDQSIYKWRGTH---TGIFHE 109
A VGDD Q+IY++ G T FHE
Sbjct 462 LFA------------------VGDDWQAIYRFSGAQMSLTTAFHE 488
> eco:b2820 recB, ECK2816, ior, JW2788, rorA; exonuclease V (RecBCD
complex), beta subunit (EC:3.1.11.5); K03582 exodeoxyribonuclease
V beta subunit [EC:3.1.11.5]
Length=1180
Score = 30.4 bits (67), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 22/98 (22%), Positives = 40/98 (40%), Gaps = 24/98 (24%)
Query 8 LDFTDLTVK---ALKLLEPPATRAELGAQWPYLVVDEFQDTNSNQFKFICLLALQRPPNI 64
L F D+ + AL+ A + ++P ++DEFQDT+ Q++ + +P
Sbjct 349 LGFDDMLSRLDSALRSESGEVLAAAIRTRFPVAMIDEFQDTDPQQYRIFRRIWHHQPET- 407
Query 65 QTLHTAVGNPTLPNSTPGRGGVTVVGDDDQSIYKWRGT 102
+ ++GD Q+IY +RG
Sbjct 408 --------------------ALLLIGDPKQAIYAFRGA 425
> dre:692126 fancb; Fanconi anemia, complementation group B; K10889
fanconi anemia group B protein
Length=807
Score = 29.6 bits (65), Expect = 2.2, Method: Composition-based stats.
Identities = 19/59 (32%), Positives = 29/59 (49%), Gaps = 2/59 (3%)
Query 53 ICLLALQRPPNIQTLHTAVGNPTLPNSTPGRGGVTVVGDDD-QSIYKWRGTHTGIFHEF 110
+C+L +RP +IQ LHT + N L + +G V V D Q + W H + +F
Sbjct 238 VCVLPYERPLDIQILHT-LKNECLIAVSFAQGHVCAVWKDTFQVVCCWSSVHLLLVDDF 295
> tpv:TP03_0243 hypothetical protein
Length=1739
Score = 29.6 bits (65), Expect = 2.6, Method: Composition-based stats.
Identities = 16/54 (29%), Positives = 25/54 (46%), Gaps = 0/54 (0%)
Query 57 ALQRPPNIQTLHTAVGNPTLPNSTPGRGGVTVVGDDDQSIYKWRGTHTGIFHEF 110
A++ P Q+ V P STP VT+ D+ QS + + G+FH +
Sbjct 47 AIKNDPVSQSTPATVTAEATPVSTPSTTPVTLDIDNTQSTSAFEYSKDGVFHSY 100
Lambda K H
0.319 0.138 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2067351240
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40