bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2356_orf1
Length=80
Score E
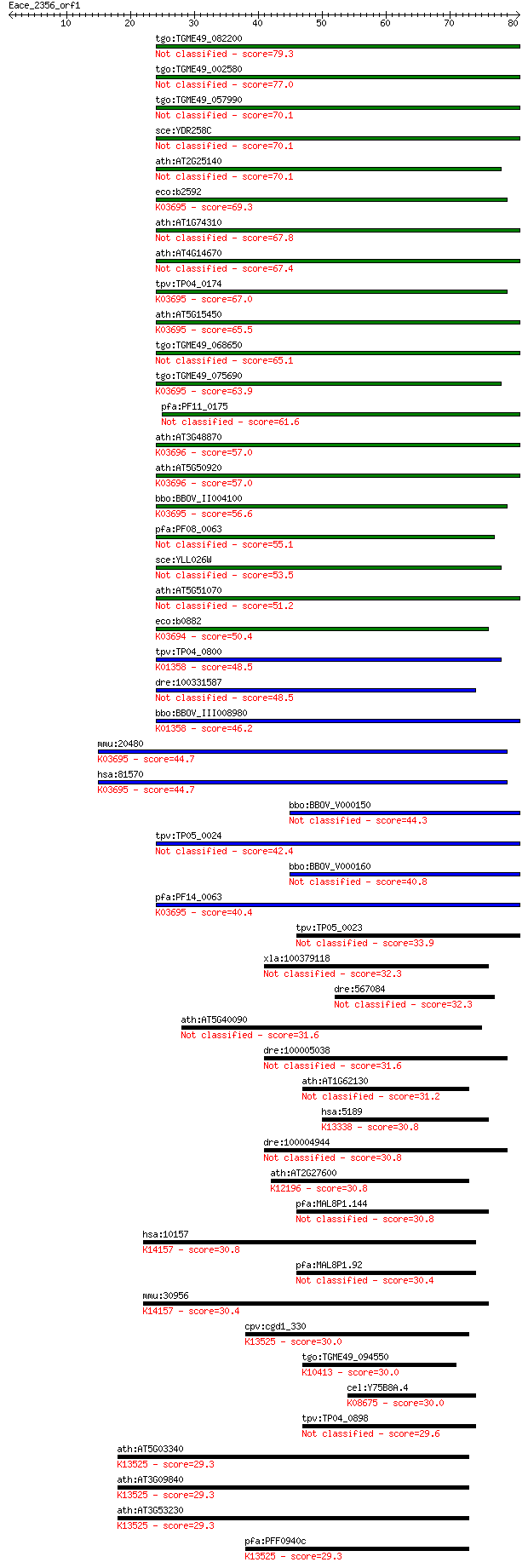
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_082200 clpB protein, putative 79.3 3e-15
tgo:TGME49_002580 heat shock protein, putative (EC:3.4.21.53) 77.0 1e-14
tgo:TGME49_057990 heat shock protein, putative (EC:3.4.21.53) 70.1 2e-12
sce:YDR258C HSP78; Hsp78p 70.1 2e-12
ath:AT2G25140 CLPB4; CLPB4 (CASEIN LYTIC PROTEINASE B4); ATP b... 70.1 2e-12
eco:b2592 clpB, ECK2590, htpM, JW2573; protein disaggregation ... 69.3 2e-12
ath:AT1G74310 ATHSP101 (ARABIDOPSIS THALIANA HEAT SHOCK PROTEI... 67.8 9e-12
ath:AT4G14670 CLPB2; CLPB2; ATP binding / nucleoside-triphosph... 67.4 1e-11
tpv:TP04_0174 hypothetical protein; K03695 ATP-dependent Clp p... 67.0 1e-11
ath:AT5G15450 CLPB3; CLPB3 (CASEIN LYTIC PROTEINASE B3); ATP b... 65.5 5e-11
tgo:TGME49_068650 clp ATP-binding chain B1, putative (EC:3.4.2... 65.1 5e-11
tgo:TGME49_075690 chaperone clpB 1 protein, putative (EC:3.4.2... 63.9 1e-10
pfa:PF11_0175 heat shock protein 101, putative 61.6 5e-10
ath:AT3G48870 HSP93-III; ATP binding / ATPase/ DNA binding / n... 57.0 1e-08
ath:AT5G50920 CLPC1; CLPC1; ATP binding / ATP-dependent peptid... 57.0 2e-08
bbo:BBOV_II004100 18.m06340; ClpB; K03695 ATP-dependent Clp pr... 56.6 2e-08
pfa:PF08_0063 ClpB protein, putative 55.1 5e-08
sce:YLL026W HSP104; Heat shock protein that cooperates with Yd... 53.5 2e-07
ath:AT5G51070 ERD1; ERD1 (EARLY RESPONSIVE TO DEHYDRATION 1); ... 51.2 7e-07
eco:b0882 clpA, ECK0873, JW0866, lopD; ATPase and specificity ... 50.4 1e-06
tpv:TP04_0800 ATP-dependent Clp protease ATP-binding subunit (... 48.5 5e-06
dre:100331587 suppressor of K+ transport defect 3-like 48.5 5e-06
bbo:BBOV_III008980 17.m07783; Clp amino terminal domain contai... 46.2 3e-05
mmu:20480 Clpb, AL118244, Skd3; ClpB caseinolytic peptidase B ... 44.7 8e-05
hsa:81570 CLPB, FLJ13152, HSP78, SKD3; ClpB caseinolytic pepti... 44.7 8e-05
bbo:BBOV_V000150 clpC 44.3 1e-04
tpv:TP05_0024 clpC; molecular chaperone 42.4 4e-04
bbo:BBOV_V000160 clpC 40.8 0.001
pfa:PF14_0063 ATP-dependent CLP protease, putative; K03695 ATP... 40.4 0.002
tpv:TP05_0023 clpC; molecular chaperone 33.9 0.12
xla:100379118 Ras-dva-2 small GTPase 32.3 0.36
dre:567084 MGC152698, MGC173525; zgc:152698 32.3 0.43
ath:AT5G40090 ATP binding / nucleoside-triphosphatase/ nucleot... 31.6 0.67
dre:100005038 hypothetical LOC100005038 31.6 0.72
ath:AT1G62130 AAA-type ATPase family protein 31.2 0.79
hsa:5189 PEX1, ZWS, ZWS1; peroxisomal biogenesis factor 1; K13... 30.8 1.0
dre:100004944 ret finger protein 2-like 30.8 1.0
ath:AT2G27600 SKD1; SKD1 (SUPPRESSOR OF K+ TRANSPORT GROWTH DE... 30.8 1.2
pfa:MAL8P1.144 AAA family ATPase, putative 30.8 1.2
hsa:10157 AASS, LKR/SDH, LKRSDH, LORSDH; aminoadipate-semialde... 30.8 1.2
pfa:MAL8P1.92 ATPase, putative 30.4 1.3
mmu:30956 Aass, LKR, LKR/SDH, LOR, LOR/SDH, Lorsdh, SDH; amino... 30.4 1.6
cpv:cgd1_330 CDC48 like AAA ATPase ortholog ; K13525 transitio... 30.0 1.9
tgo:TGME49_094550 dynein heavy chain, putative ; K10413 dynein... 30.0 2.0
cel:Y75B8A.4 hypothetical protein; K08675 Lon-like ATP-depende... 30.0 2.1
tpv:TP04_0898 hypothetical protein 29.6 2.7
ath:AT5G03340 cell division cycle protein 48, putative / CDC48... 29.3 3.1
ath:AT3G09840 CDC48; CDC48 (CELL DIVISION CYCLE 48); ATPase/ i... 29.3 3.1
ath:AT3G53230 cell division cycle protein 48, putative / CDC48... 29.3 3.4
pfa:PFF0940c cell division cycle protein 48 homologue, putativ... 29.3 3.5
> tgo:TGME49_082200 clpB protein, putative
Length=970
Score = 79.3 bits (194), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 39/57 (68%), Positives = 46/57 (80%), Gaps = 0/57 (0%)
Query 24 QQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSEKNL 80
Q AVKAVADA+ RAGLS + P+G+F+FLG +GVGKTELAKA+A EMF SEKNL
Sbjct 590 QDDAVKAVADAMVRARAGLSREGMPVGSFLFLGPTGVGKTELAKALAMEMFHSEKNL 646
> tgo:TGME49_002580 heat shock protein, putative (EC:3.4.21.53)
Length=983
Score = 77.0 bits (188), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 35/57 (61%), Positives = 47/57 (82%), Gaps = 0/57 (0%)
Query 24 QQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSEKNL 80
Q++AV+AVA+A+ RAGL+ +N P+GTF+FLGS+GVGKTELAKA+ +F EKNL
Sbjct 603 QEEAVEAVANAIIRSRAGLARRNAPIGTFLFLGSTGVGKTELAKALTAAIFHDEKNL 659
> tgo:TGME49_057990 heat shock protein, putative (EC:3.4.21.53)
Length=921
Score = 70.1 bits (170), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 33/57 (57%), Positives = 44/57 (77%), Gaps = 0/57 (0%)
Query 24 QQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSEKNL 80
Q QAV+AV A+ AGLS +N+P+G+F+FLG +GVGKTEL K VAE +FDS++ L
Sbjct 575 QPQAVEAVTQAILRSAAGLSRRNRPIGSFLFLGPTGVGKTELCKRVAESLFDSKERL 631
> sce:YDR258C HSP78; Hsp78p
Length=811
Score = 70.1 bits (170), Expect = 2e-12, Method: Composition-based stats.
Identities = 30/57 (52%), Positives = 45/57 (78%), Gaps = 0/57 (0%)
Query 24 QQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSEKNL 80
Q +A+ A++DA+ +QRAGL+ + +P+ +FMFLG +G GKTEL KA+AE +FD E N+
Sbjct 509 QDEAIAAISDAVRLQRAGLTSEKRPIASFMFLGPTGTGKTELTKALAEFLFDDESNV 565
> ath:AT2G25140 CLPB4; CLPB4 (CASEIN LYTIC PROTEINASE B4); ATP
binding / ATPase/ nucleoside-triphosphatase/ nucleotide binding
/ protein binding
Length=964
Score = 70.1 bits (170), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 32/54 (59%), Positives = 43/54 (79%), Gaps = 0/54 (0%)
Query 24 QQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSE 77
Q AVK+VADA+ RAGLS N+P+ +FMF+G +GVGKTELAKA+A +F++E
Sbjct 658 QDMAVKSVADAIRRSRAGLSDPNRPIASFMFMGPTGVGKTELAKALAGYLFNTE 711
> eco:b2592 clpB, ECK2590, htpM, JW2573; protein disaggregation
chaperone; K03695 ATP-dependent Clp protease ATP-binding subunit
ClpB
Length=857
Score = 69.3 bits (168), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 31/55 (56%), Positives = 44/55 (80%), Gaps = 0/55 (0%)
Query 24 QQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSEK 78
Q +AV AV++A+ RAGL+ N+P+G+F+FLG +GVGKTEL KA+A MFDS++
Sbjct 573 QNEAVDAVSNAIRRSRAGLADPNRPIGSFLFLGPTGVGKTELCKALANFMFDSDE 627
> ath:AT1G74310 ATHSP101 (ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN
101); ATP binding / ATPase/ nucleoside-triphosphatase/ nucleotide
binding / protein binding
Length=911
Score = 67.8 bits (164), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 33/57 (57%), Positives = 43/57 (75%), Gaps = 0/57 (0%)
Query 24 QQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSEKNL 80
Q QAV AV++A+ RAGL +P G+F+FLG +GVGKTELAKA+AE++FD E L
Sbjct 574 QNQAVNAVSEAILRSRAGLGRPQQPTGSFLFLGPTGVGKTELAKALAEQLFDDENLL 630
> ath:AT4G14670 CLPB2; CLPB2; ATP binding / nucleoside-triphosphatase/
nucleotide binding / protein binding
Length=623
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 34/57 (59%), Positives = 43/57 (75%), Gaps = 0/57 (0%)
Query 24 QQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSEKNL 80
Q +AVKAVA A+ R GL +P G+F+FLG +GVGKTELAKA+AE++FDSE L
Sbjct 539 QDEAVKAVAAAILRSRVGLGRPQQPSGSFLFLGPTGVGKTELAKALAEQLFDSENLL 595
> tpv:TP04_0174 hypothetical protein; K03695 ATP-dependent Clp
protease ATP-binding subunit ClpB
Length=985
Score = 67.0 bits (162), Expect = 1e-11, Method: Composition-based stats.
Identities = 29/55 (52%), Positives = 42/55 (76%), Gaps = 0/55 (0%)
Query 24 QQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSEK 78
QQ+A+ AV +A+ R G++ KP+ MFLG +GVGKTEL+KA+AE++FDSE+
Sbjct 699 QQEAIDAVVNAVQRSRVGMNDPKKPIAALMFLGPTGVGKTELSKAIAEQLFDSEE 753
> ath:AT5G15450 CLPB3; CLPB3 (CASEIN LYTIC PROTEINASE B3); ATP
binding / ATPase/ nucleoside-triphosphatase/ nucleotide binding
/ protein binding; K03695 ATP-dependent Clp protease ATP-binding
subunit ClpB
Length=968
Score = 65.5 bits (158), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 32/57 (56%), Positives = 43/57 (75%), Gaps = 0/57 (0%)
Query 24 QQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSEKNL 80
Q AV AVA+A+ RAGLS +P+ +FMF+G +GVGKTELAKA+A MF++E+ L
Sbjct 653 QNPAVTAVAEAIQRSRAGLSDPGRPIASFMFMGPTGVGKTELAKALASYMFNTEEAL 709
> tgo:TGME49_068650 clp ATP-binding chain B1, putative (EC:3.4.21.53)
Length=929
Score = 65.1 bits (157), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 28/57 (49%), Positives = 44/57 (77%), Gaps = 0/57 (0%)
Query 24 QQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSEKNL 80
Q++ V++VA+A+ RAGL N+P+ + +FLG +GVGKTEL KA+A ++FD+E+ L
Sbjct 599 QEEGVRSVAEAIQRSRAGLCDPNRPIASLVFLGPTGVGKTELCKALARQLFDTEEAL 655
> tgo:TGME49_075690 chaperone clpB 1 protein, putative (EC:3.4.21.53);
K03695 ATP-dependent Clp protease ATP-binding subunit
ClpB
Length=898
Score = 63.9 bits (154), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 29/54 (53%), Positives = 40/54 (74%), Gaps = 0/54 (0%)
Query 24 QQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSE 77
Q AV+ VA+A+ RAGL+ N+P+ + FLG +GVGKTEL +++AE MFDSE
Sbjct 731 QDHAVQVVAEAIQRSRAGLNDPNRPIASLFFLGPTGVGKTELCRSLAELMFDSE 784
> pfa:PF11_0175 heat shock protein 101, putative
Length=906
Score = 61.6 bits (148), Expect = 5e-10, Method: Composition-based stats.
Identities = 27/56 (48%), Positives = 41/56 (73%), Gaps = 0/56 (0%)
Query 25 QQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSEKNL 80
+ +K+++DA+ G+ KP+GTF+FLG +GVGKTELAK +A E+F+S+ NL
Sbjct 607 EDIIKSLSDAVVKAATGMKDPEKPIGTFLFLGPTGVGKTELAKTLAIELFNSKDNL 662
> ath:AT3G48870 HSP93-III; ATP binding / ATPase/ DNA binding /
nuclease/ nucleoside-triphosphatase/ nucleotide binding / protein
binding; K03696 ATP-dependent Clp protease ATP-binding
subunit ClpC
Length=952
Score = 57.0 bits (136), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 27/57 (47%), Positives = 40/57 (70%), Gaps = 0/57 (0%)
Query 24 QQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSEKNL 80
Q +AVKA++ A+ R GL N+P+ +F+F G +GVGK+ELAKA+A F SE+ +
Sbjct 634 QDEAVKAISRAIRRARVGLKNPNRPIASFIFSGPTGVGKSELAKALAAYYFGSEEAM 690
> ath:AT5G50920 CLPC1; CLPC1; ATP binding / ATP-dependent peptidase/
ATPase; K03696 ATP-dependent Clp protease ATP-binding
subunit ClpC
Length=929
Score = 57.0 bits (136), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 27/57 (47%), Positives = 40/57 (70%), Gaps = 0/57 (0%)
Query 24 QQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSEKNL 80
Q +AVKA++ A+ R GL N+P+ +F+F G +GVGK+ELAKA+A F SE+ +
Sbjct 613 QDEAVKAISRAIRRARVGLKNPNRPIASFIFSGPTGVGKSELAKALAAYYFGSEEAM 669
> bbo:BBOV_II004100 18.m06340; ClpB; K03695 ATP-dependent Clp
protease ATP-binding subunit ClpB
Length=931
Score = 56.6 bits (135), Expect = 2e-08, Method: Composition-based stats.
Identities = 25/55 (45%), Positives = 38/55 (69%), Gaps = 0/55 (0%)
Query 24 QQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSEK 78
Q +AV V A+ R G++ +P+ MFLG +GVGKTEL KA+AE++FD+++
Sbjct 644 QDEAVDIVTRAVQRSRVGMNDPKRPIAGLMFLGPTGVGKTELCKAIAEQLFDTDE 698
> pfa:PF08_0063 ClpB protein, putative
Length=1070
Score = 55.1 bits (131), Expect = 5e-08, Method: Composition-based stats.
Identities = 24/53 (45%), Positives = 36/53 (67%), Gaps = 0/53 (0%)
Query 24 QQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFDS 76
Q AVK V A+ R G++ +P+ + MFLG +GVGKTEL+K +A+ +FD+
Sbjct 784 QDDAVKVVTKAVQRSRVGMNNPKRPIASLMFLGPTGVGKTELSKVLADVLFDT 836
> sce:YLL026W HSP104; Heat shock protein that cooperates with
Ydj1p (Hsp40) and Ssa1p (Hsp70) to refold and reactivate previously
denatured, aggregated proteins; responsive to stresses
including: heat, ethanol, and sodium arsenite; involved in
[PSI+] propagation
Length=908
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 27/54 (50%), Positives = 39/54 (72%), Gaps = 1/54 (1%)
Query 24 QQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSE 77
Q A+KAV++A+ + R+GL+ +P +F+FLG SG GKTELAK VA +F+ E
Sbjct 583 QMDAIKAVSNAVRLSRSGLANPRQP-ASFLFLGLSGSGKTELAKKVAGFLFNDE 635
> ath:AT5G51070 ERD1; ERD1 (EARLY RESPONSIVE TO DEHYDRATION 1);
ATP binding / ATPase/ nucleoside-triphosphatase/ nucleotide
binding / protein binding
Length=945
Score = 51.2 bits (121), Expect = 7e-07, Method: Composition-based stats.
Identities = 24/57 (42%), Positives = 37/57 (64%), Gaps = 0/57 (0%)
Query 24 QQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSEKNL 80
Q +AV A++ A+ R GL ++P+ +F G +GVGKTEL KA+A F SE+++
Sbjct 632 QDEAVAAISRAVKRSRVGLKDPDRPIAAMLFCGPTGVGKTELTKALAANYFGSEESM 688
> eco:b0882 clpA, ECK0873, JW0866, lopD; ATPase and specificity
subunit of ClpA-ClpP ATP-dependent serine protease, chaperone
activity; K03694 ATP-dependent Clp protease ATP-binding
subunit ClpA
Length=758
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 26/58 (44%), Positives = 42/58 (72%), Gaps = 6/58 (10%)
Query 24 QQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTE----LAKAVAEEM--FD 75
Q +A++A+ +A+ + RAGL ++KP+G+F+F G +GVGKTE L+KA+ E+ FD
Sbjct 463 QDKAIEALTEAIKMARAGLGHEHKPVGSFLFAGPTGVGKTEVTVQLSKALGIELLRFD 520
> tpv:TP04_0800 ATP-dependent Clp protease ATP-binding subunit
(EC:3.4.21.92); K01358 ATP-dependent Clp protease, protease
subunit [EC:3.4.21.92]
Length=900
Score = 48.5 bits (114), Expect = 5e-06, Method: Composition-based stats.
Identities = 21/54 (38%), Positives = 36/54 (66%), Gaps = 0/54 (0%)
Query 24 QQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSE 77
Q++AVK V A+ + + N+P+G+F+F G GVGK+E+A+A+ + +F E
Sbjct 578 QEEAVKNVCKAIRRAKTNIKNPNRPIGSFLFCGPPGVGKSEVARALTKYLFAKE 631
> dre:100331587 suppressor of K+ transport defect 3-like
Length=409
Score = 48.5 bits (114), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 25/50 (50%), Positives = 32/50 (64%), Gaps = 1/50 (2%)
Query 24 QQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEM 73
Q+ A+ VA A+ + G + PL F+FLGSSG+GKTELAK VA M
Sbjct 61 QEGAINTVASAIRRKENGWYDEEHPL-VFLFLGSSGIGKTELAKQVARYM 109
> bbo:BBOV_III008980 17.m07783; Clp amino terminal domain containing
protein; K01358 ATP-dependent Clp protease, protease
subunit [EC:3.4.21.92]
Length=1005
Score = 46.2 bits (108), Expect = 3e-05, Method: Composition-based stats.
Identities = 22/57 (38%), Positives = 37/57 (64%), Gaps = 1/57 (1%)
Query 24 QQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSEKNL 80
++AVK +A A+ + + +P+G+F+F G GVGK+E+AK + + MF +E NL
Sbjct 664 HEEAVKNIAKAIRRAKTNIKNPERPIGSFLFCGPPGVGKSEIAKTLTKLMF-TEDNL 719
> mmu:20480 Clpb, AL118244, Skd3; ClpB caseinolytic peptidase
B homolog (E. coli); K03695 ATP-dependent Clp protease ATP-binding
subunit ClpB
Length=677
Score = 44.7 bits (104), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 27/67 (40%), Positives = 37/67 (55%), Gaps = 4/67 (5%)
Query 15 PQQQQLPRQ---QQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAE 71
P +Q+L Q+ A+ V A+ + G + PL F+FLGSSG+GKTELAK A+
Sbjct 308 PLEQRLKEHIIGQESAIATVGAAIRRKENGWYDEEHPL-VFLFLGSSGIGKTELAKQTAK 366
Query 72 EMFDSEK 78
M K
Sbjct 367 YMHKDAK 373
> hsa:81570 CLPB, FLJ13152, HSP78, SKD3; ClpB caseinolytic peptidase
B homolog (E. coli); K03695 ATP-dependent Clp protease
ATP-binding subunit ClpB
Length=707
Score = 44.7 bits (104), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 27/67 (40%), Positives = 37/67 (55%), Gaps = 4/67 (5%)
Query 15 PQQQQLPRQ---QQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAE 71
P +Q+L Q+ A+ V A+ + G + PL F+FLGSSG+GKTELAK A+
Sbjct 338 PLEQRLKEHIIGQESAIATVGAAIRRKENGWYDEEHPL-VFLFLGSSGIGKTELAKQTAK 396
Query 72 EMFDSEK 78
M K
Sbjct 397 YMHKDAK 403
> bbo:BBOV_V000150 clpC
Length=541
Score = 44.3 bits (103), Expect = 1e-04, Method: Composition-based stats.
Identities = 20/36 (55%), Positives = 23/36 (63%), Gaps = 0/36 (0%)
Query 45 KNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSEKNL 80
K KPL +F+F G SG GKTELAK +F S K L
Sbjct 274 KTKPLSSFLFCGPSGAGKTELAKIFTYSLFKSTKQL 309
> tpv:TP05_0024 clpC; molecular chaperone
Length=529
Score = 42.4 bits (98), Expect = 4e-04, Method: Composition-based stats.
Identities = 22/57 (38%), Positives = 33/57 (57%), Gaps = 0/57 (0%)
Query 24 QQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSEKNL 80
Q +AV++V + + KPLG+++ G SG GKTELAK + +F+S NL
Sbjct 246 QDEAVESVLYRIKKLSNANIDETKPLGSWLLCGPSGTGKTELAKILCYTIFNSHDNL 302
> bbo:BBOV_V000160 clpC
Length=551
Score = 40.8 bits (94), Expect = 0.001, Method: Composition-based stats.
Identities = 16/36 (44%), Positives = 24/36 (66%), Gaps = 0/36 (0%)
Query 45 KNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSEKNL 80
K KP+G+F+ G SG GKTE+ K + ++ S+ NL
Sbjct 299 KIKPIGSFLLCGPSGTGKTEIVKLITNYLYKSQTNL 334
> pfa:PF14_0063 ATP-dependent CLP protease, putative; K03695 ATP-dependent
Clp protease ATP-binding subunit ClpB
Length=1341
Score = 40.4 bits (93), Expect = 0.002, Method: Composition-based stats.
Identities = 20/57 (35%), Positives = 34/57 (59%), Gaps = 1/57 (1%)
Query 24 QQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSEKNL 80
Q++ + ++ L + NKP+GT + GSSGVGKT A+ +++ +F+ E NL
Sbjct 936 QEKVIDILSKYLFKAITNIKDPNKPIGTLLLCGSSGVGKTLCAQVISKYLFN-EDNL 991
> tpv:TP05_0023 clpC; molecular chaperone
Length=502
Score = 33.9 bits (76), Expect = 0.12, Method: Composition-based stats.
Identities = 13/35 (37%), Positives = 22/35 (62%), Gaps = 0/35 (0%)
Query 46 NKPLGTFMFLGSSGVGKTELAKAVAEEMFDSEKNL 80
NKP+ F+ G SG GKT++ K ++ + ++NL
Sbjct 255 NKPIANFLLCGPSGTGKTDICKILSTYLGSEKQNL 289
> xla:100379118 Ras-dva-2 small GTPase
Length=209
Score = 32.3 bits (72), Expect = 0.36, Method: Compositional matrix adjust.
Identities = 15/35 (42%), Positives = 21/35 (60%), Gaps = 0/35 (0%)
Query 41 GLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFD 75
LS K K +FLG++GVGKT L + ++ FD
Sbjct 2 SLSTKEKRQIRLVFLGAAGVGKTSLIRRFLQDTFD 36
> dre:567084 MGC152698, MGC173525; zgc:152698
Length=211
Score = 32.3 bits (72), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 13/25 (52%), Positives = 18/25 (72%), Gaps = 0/25 (0%)
Query 52 FMFLGSSGVGKTELAKAVAEEMFDS 76
F+FLG++GVGKT L ++ FDS
Sbjct 13 FVFLGAAGVGKTALITRFLQDRFDS 37
> ath:AT5G40090 ATP binding / nucleoside-triphosphatase/ nucleotide
binding / transmembrane receptor
Length=459
Score = 31.6 bits (70), Expect = 0.67, Method: Composition-based stats.
Identities = 19/47 (40%), Positives = 27/47 (57%), Gaps = 6/47 (12%)
Query 28 VKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMF 74
+K V D LA++ NK + T GS+GVGKT LA+ + E+F
Sbjct 188 MKVVYDLLALE------VNKEVRTIGIWGSAGVGKTTLARYIYAEIF 228
> dre:100005038 hypothetical LOC100005038
Length=498
Score = 31.6 bits (70), Expect = 0.72, Method: Composition-based stats.
Identities = 14/38 (36%), Positives = 20/38 (52%), Gaps = 0/38 (0%)
Query 41 GLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSEK 78
G KNKP T + +G +G GKT+L + M E+
Sbjct 50 GKRDKNKPHKTILMVGETGTGKTKLINTMVNYMLGVER 87
> ath:AT1G62130 AAA-type ATPase family protein
Length=1025
Score = 31.2 bits (69), Expect = 0.79, Method: Composition-based stats.
Identities = 15/26 (57%), Positives = 16/26 (61%), Gaps = 0/26 (0%)
Query 47 KPLGTFMFLGSSGVGKTELAKAVAEE 72
KP + G SG GKT LAKAVA E
Sbjct 768 KPCNGILLFGPSGTGKTMLAKAVATE 793
> hsa:5189 PEX1, ZWS, ZWS1; peroxisomal biogenesis factor 1; K13338
peroxin-1
Length=1283
Score = 30.8 bits (68), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 12/26 (46%), Positives = 16/26 (61%), Gaps = 0/26 (0%)
Query 50 GTFMFLGSSGVGKTELAKAVAEEMFD 75
G + G G GK+ LAKA+ +E FD
Sbjct 593 GALLLTGGKGSGKSTLAKAICKEAFD 618
> dre:100004944 ret finger protein 2-like
Length=600
Score = 30.8 bits (68), Expect = 1.0, Method: Composition-based stats.
Identities = 14/38 (36%), Positives = 20/38 (52%), Gaps = 0/38 (0%)
Query 41 GLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSEK 78
G +NKP T + +G +GVGKT L + M E+
Sbjct 131 GKRDENKPHKTILIVGETGVGKTTLVNVMVNYMLGVER 168
> ath:AT2G27600 SKD1; SKD1 (SUPPRESSOR OF K+ TRANSPORT GROWTH
DEFECT1); ATP binding / nucleoside-triphosphatase/ nucleotide
binding; K12196 vacuolar protein-sorting-associated protein
4
Length=435
Score = 30.8 bits (68), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 14/31 (45%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 42 LSPKNKPLGTFMFLGSSGVGKTELAKAVAEE 72
+ K +P F+ G G GK+ LAKAVA E
Sbjct 158 FTGKRRPWRAFLLYGPPGTGKSYLAKAVATE 188
> pfa:MAL8P1.144 AAA family ATPase, putative
Length=1467
Score = 30.8 bits (68), Expect = 1.2, Method: Composition-based stats.
Identities = 16/33 (48%), Positives = 22/33 (66%), Gaps = 3/33 (9%)
Query 46 NKPLG--TFMFLGSSGVGKTELAKAVAEEM-FD 75
N+P T +F G +G GKT LAK +A+E+ FD
Sbjct 691 NEPFSCDTILFSGDTGTGKTMLAKTMAKELNFD 723
> hsa:10157 AASS, LKR/SDH, LKRSDH, LORSDH; aminoadipate-semialdehyde
synthase (EC:1.5.1.8 1.5.1.9); K14157 alpha-aminoadipic
semialdehyde synthase [EC:1.5.1.8 1.5.1.9]
Length=926
Score = 30.8 bits (68), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 21/52 (40%), Positives = 28/52 (53%), Gaps = 2/52 (3%)
Query 22 RQQQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEM 73
R QAV+AV DA GL PK+ TF+F G+ V K A+A+ E+
Sbjct 189 RNSSQAVQAVRDAGYEISLGLMPKSIGPLTFVFTGTGNVSKG--AQAIFNEL 238
> pfa:MAL8P1.92 ATPase, putative
Length=1200
Score = 30.4 bits (67), Expect = 1.3, Method: Composition-based stats.
Identities = 14/28 (50%), Positives = 19/28 (67%), Gaps = 1/28 (3%)
Query 46 NKPLGTFMFLGSSGVGKTELAKAVAEEM 73
NK +G ++ G G GKT LAKA++ EM
Sbjct 708 NKSMGILLY-GPPGCGKTMLAKAISNEM 734
> mmu:30956 Aass, LKR, LKR/SDH, LOR, LOR/SDH, Lorsdh, SDH; aminoadipate-semialdehyde
synthase (EC:1.5.1.8 1.5.1.9); K14157
alpha-aminoadipic semialdehyde synthase [EC:1.5.1.8 1.5.1.9]
Length=926
Score = 30.4 bits (67), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 21/54 (38%), Positives = 28/54 (51%), Gaps = 6/54 (11%)
Query 22 RQQQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFD 75
R QAV+AV DA GL PK+ TF+F G+ V K A+E+F+
Sbjct 189 RNSSQAVQAVRDAGYEISLGLMPKSIGPLTFVFTGTGNVSKG------AQEVFN 236
> cpv:cgd1_330 CDC48 like AAA ATPase ortholog ; K13525 transitional
endoplasmic reticulum ATPase
Length=820
Score = 30.0 bits (66), Expect = 1.9, Method: Composition-based stats.
Identities = 18/35 (51%), Positives = 20/35 (57%), Gaps = 4/35 (11%)
Query 38 QRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEE 72
+R G+SP L F G G GKT LAKAVA E
Sbjct 524 ERFGMSPSRGVL----FYGPPGCGKTLLAKAVASE 554
> tgo:TGME49_094550 dynein heavy chain, putative ; K10413 dynein
heavy chain 1, cytosolic
Length=4937
Score = 30.0 bits (66), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 13/24 (54%), Positives = 17/24 (70%), Gaps = 0/24 (0%)
Query 47 KPLGTFMFLGSSGVGKTELAKAVA 70
+PLG + +G+SG GKT L K VA
Sbjct 3058 QPLGHLLLVGASGAGKTILTKFVA 3081
> cel:Y75B8A.4 hypothetical protein; K08675 Lon-like ATP-dependent
protease [EC:3.4.21.-]
Length=773
Score = 30.0 bits (66), Expect = 2.1, Method: Composition-based stats.
Identities = 12/20 (60%), Positives = 15/20 (75%), Gaps = 0/20 (0%)
Query 54 FLGSSGVGKTELAKAVAEEM 73
F G G+GKT +AKA+AE M
Sbjct 334 FTGPPGIGKTSIAKAIAESM 353
> tpv:TP04_0898 hypothetical protein
Length=791
Score = 29.6 bits (65), Expect = 2.7, Method: Composition-based stats.
Identities = 13/27 (48%), Positives = 18/27 (66%), Gaps = 0/27 (0%)
Query 47 KPLGTFMFLGSSGVGKTELAKAVAEEM 73
+P + G SGVGKT LA+ +A+EM
Sbjct 549 RPSVGLVIKGDSGVGKTSLAQCLAQEM 575
> ath:AT5G03340 cell division cycle protein 48, putative / CDC48,
putative; K13525 transitional endoplasmic reticulum ATPase
Length=810
Score = 29.3 bits (64), Expect = 3.1, Method: Composition-based stats.
Identities = 20/56 (35%), Positives = 29/56 (51%), Gaps = 5/56 (8%)
Query 18 QQLPRQQQQAVK-AVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEE 72
+ + R+ Q+ V+ V ++ G+SP L F G G GKT LAKA+A E
Sbjct 486 ENVKRELQETVQYPVEHPEKFEKFGMSPSKGVL----FYGPPGCGKTLLAKAIANE 537
> ath:AT3G09840 CDC48; CDC48 (CELL DIVISION CYCLE 48); ATPase/
identical protein binding; K13525 transitional endoplasmic
reticulum ATPase
Length=809
Score = 29.3 bits (64), Expect = 3.1, Method: Composition-based stats.
Identities = 20/56 (35%), Positives = 29/56 (51%), Gaps = 5/56 (8%)
Query 18 QQLPRQQQQAVK-AVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEE 72
+ + R+ Q+ V+ V ++ G+SP L F G G GKT LAKA+A E
Sbjct 486 ENVKRELQETVQYPVEHPEKFEKFGMSPSKGVL----FYGPPGCGKTLLAKAIANE 537
> ath:AT3G53230 cell division cycle protein 48, putative / CDC48,
putative; K13525 transitional endoplasmic reticulum ATPase
Length=815
Score = 29.3 bits (64), Expect = 3.4, Method: Composition-based stats.
Identities = 20/56 (35%), Positives = 29/56 (51%), Gaps = 5/56 (8%)
Query 18 QQLPRQQQQAVK-AVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEE 72
+ + R+ Q+ V+ V ++ G+SP L F G G GKT LAKA+A E
Sbjct 487 ENVKRELQETVQYPVEHPEKFEKFGMSPSKGVL----FYGPPGCGKTLLAKAIANE 538
> pfa:PFF0940c cell division cycle protein 48 homologue, putative;
K13525 transitional endoplasmic reticulum ATPase
Length=828
Score = 29.3 bits (64), Expect = 3.5, Method: Composition-based stats.
Identities = 17/35 (48%), Positives = 20/35 (57%), Gaps = 4/35 (11%)
Query 38 QRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEE 72
++ G+SP L F G G GKT LAKAVA E
Sbjct 507 EKFGMSPSRGVL----FYGPPGCGKTLLAKAVASE 537
Lambda K H
0.312 0.127 0.357
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2058492688
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40