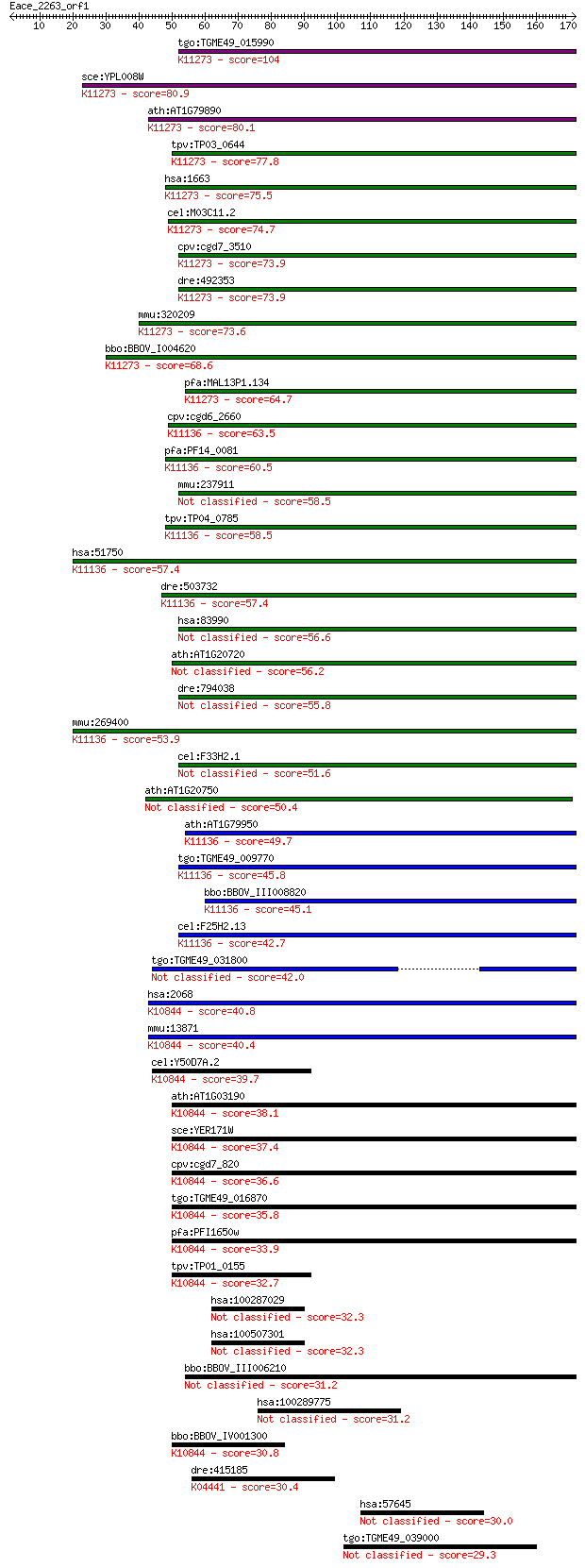
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2263_orf1
Length=171
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_015990 helicase, putative ; K11273 chromosome trans... 104 1e-22
sce:YPL008W CHL1, CTF1, LPA9, MCM12; Chl1p (EC:3.6.1.-); K1127... 80.9 2e-15
ath:AT1G79890 helicase-related; K11273 chromosome transmission... 80.1 3e-15
tpv:TP03_0644 DNA helicase; K11273 chromosome transmission fid... 77.8 2e-14
hsa:1663 DDX11, CHL1, CHLR1, KRG2, MGC133249, MGC9335; DEAD/H ... 75.5 7e-14
cel:M03C11.2 hypothetical protein; K11273 chromosome transmiss... 74.7 2e-13
cpv:cgd7_3510 helicase ; K11273 chromosome transmission fideli... 73.9 2e-13
dre:492353 zgc:92172 (EC:3.6.4.13); K11273 chromosome transmis... 73.9 3e-13
mmu:320209 Ddx11, 4732462I11Rik, CHL1, CHLR1, KRG2, MGC90809; ... 73.6 3e-13
bbo:BBOV_I004620 19.m02044; DNA repair helicase (rad3) and DEA... 68.6 9e-12
pfa:MAL13P1.134 DEAD box helicase, putative; K11273 chromosome... 64.7 1e-10
cpv:cgd6_2660 DNA repair helicase ; K11136 regulator of telome... 63.5 3e-10
pfa:PF14_0081 DNA-repair helicase, putative; K11136 regulator ... 60.5 2e-09
mmu:237911 Brip1, 3110009N10Rik, 8030460J03Rik, Bach1, FACJ, F... 58.5 9e-09
tpv:TP04_0785 DNA repair helicase; K11136 regulator of telomer... 58.5 1e-08
hsa:51750 RTEL1, C20orf41, DKFZp434C013, KIAA1088, NHL, RTEL, ... 57.4 2e-08
dre:503732 rtel1; zgc:113114 (EC:3.6.4.12); K11136 regulator o... 57.4 2e-08
hsa:83990 BRIP1, BACH1, FANCJ, FLJ90232, MGC126521, MGC126523,... 56.6 4e-08
ath:AT1G20720 ATP binding / ATP-dependent DNA helicase/ ATP-de... 56.2 6e-08
dre:794038 brip1, fancj, si:ch211-158l18.1; BRCA1 interacting ... 55.8 7e-08
mmu:269400 Rtel1, AI451565, AW540478, Rtel; regulator of telom... 53.9 2e-07
cel:F33H2.1 dog-1; Deletions Of G-rich DNA family member (dog-1) 51.6 1e-06
ath:AT1G20750 helicase-related 50.4 3e-06
ath:AT1G79950 helicase-related; K11136 regulator of telomere e... 49.7 5e-06
tgo:TGME49_009770 DNA repair helicase, putative ; K11136 regul... 45.8 8e-05
bbo:BBOV_III008820 17.m07769; DNA repair helicase (rad3) famil... 45.1 1e-04
cel:F25H2.13 rtel-1; RTEL (mammalian Regulator of TELomere len... 42.7 6e-04
tgo:TGME49_031800 hypothetical protein 42.0 0.001
hsa:2068 ERCC2, COFS2, EM9, MGC102762, MGC126218, MGC126219, T... 40.8 0.002
mmu:13871 Ercc2, AA407812, AU020867, AW240756, Ercc-2, XPD; ex... 40.4 0.003
cel:Y50D7A.2 hypothetical protein; K10844 DNA excision repair ... 39.7 0.004
ath:AT1G03190 UVH6; UVH6 (ULTRAVIOLET HYPERSENSITIVE 6); ATP b... 38.1 0.016
sce:YER171W RAD3, REM1; 5' to 3' DNA helicase, involved in nuc... 37.4 0.023
cpv:cgd7_820 RAD3'DEXDc+HELICc protein' ; K10844 DNA excision ... 36.6 0.047
tgo:TGME49_016870 excision repair protein rad15, putative ; K1... 35.8 0.069
pfa:PFI1650w DNA excision-repair helicase, putative; K10844 DN... 33.9 0.29
tpv:TP01_0155 DNA repair protein Rad3; K10844 DNA excision rep... 32.7 0.55
hsa:100287029 DDX11L10, DDX11L1, DDX11P; DEAD/H (Asp-Glu-Ala-A... 32.3 0.72
hsa:100507301 hypothetical protein LOC100507301 32.3 0.75
bbo:BBOV_III006210 17.m07551; hypothetical protein 31.2 1.6
hsa:100289775 hypothetical protein LOC100289775 31.2 1.8
bbo:BBOV_IV001300 21.m02831; DNA excision repair helicase; K10... 30.8 2.2
dre:415185 zgc:86905; K04441 p38 MAP kinase [EC:2.7.11.24] 30.4 3.0
hsa:57645 POGK, BASS2, KIAA1513, KIAA15131, KRBOX2; pogo trans... 30.0 3.6
tgo:TGME49_039000 hypothetical protein 29.3 7.2
> tgo:TGME49_015990 helicase, putative ; K11273 chromosome transmission
fidelity protein 1 [EC:3.6.4.13]
Length=1282
Score = 104 bits (260), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 65/167 (38%), Positives = 82/167 (49%), Gaps = 47/167 (28%)
Query 52 AFLFCVMGGRLSEGVNFSNALARLLVVVGMPFPSVKDKIFILHKEHYNAIMTNPKATAD- 110
A LFCVM G LSEGVNF ++LARLLV+VG+PFPS+KD F L +++++ + AD
Sbjct 1019 AALFCVMNGALSEGVNFHDSLARLLVLVGLPFPSIKDPEFQLRAAFFSSVLERTQRNADT 1078
Query 111 -----EAEEAKPAPSDWPAD----------AKQLQQQQREQH------------------ 137
E E+ + D A Q Q+ H
Sbjct 1079 HASVGEKRESDTTRVEKNVDKTDSGAARFRAAQTTAQENSVHGRGGNRSMDAEASTVRKL 1138
Query 138 -------------FVDYGLLQCMITVNQTIGRAIRHSKDYAAILLVD 171
YGLL C+ TVNQTIGRAIRHS+DYAA+LLVD
Sbjct 1139 ERKPRCAEPTPNGRTSYGLLHCLQTVNQTIGRAIRHSQDYAAVLLVD 1185
> sce:YPL008W CHL1, CTF1, LPA9, MCM12; Chl1p (EC:3.6.1.-); K11273
chromosome transmission fidelity protein 1 [EC:3.6.4.13]
Length=861
Score = 80.9 bits (198), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 47/149 (31%), Positives = 75/149 (50%), Gaps = 22/149 (14%)
Query 23 NNRSKRGFQWEDKTCKALSAQESSPQSKTAFLFCVMGGRLSEGVNFSNALARLLVVVGMP 82
NN K ++ +D +S + + + L ++GG+LSEG+NF + L R +V+VG+P
Sbjct 690 NNVRKIFYEAKDGDDILSGYSDSVAEGRGSLLLAIVGGKLSEGINFQDDLCRAVVMVGLP 749
Query 83 FPSVKDKIFILHKEHYNAIMTNPKATADEAEEAKPAPSDWPADAKQLQQQQREQHFVDYG 142
FP++ I+ ++H A + T +EA A K+ +
Sbjct 750 FPNIFSGELIVKRKHLAAKIMKSGGTEEEASRA----------TKEFMEN---------- 789
Query 143 LLQCMITVNQTIGRAIRHSKDYAAILLVD 171
CM VNQ++GRAIRH+ DYA I L+D
Sbjct 790 --ICMKAVNQSVGRAIRHANDYANIYLLD 816
> ath:AT1G79890 helicase-related; K11273 chromosome transmission
fidelity protein 1 [EC:3.6.4.13]
Length=882
Score = 80.1 bits (196), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 48/135 (35%), Positives = 76/135 (56%), Gaps = 13/135 (9%)
Query 43 QESSPQSKTAFLFCVMGGRLSEGVNFSNALARLLVVVGMPFPSVKDKIFILHKEHYNAIM 102
+E+ + A + V+GG++SEG+NFS+++ R +V+VG+P+PS D + +H
Sbjct 695 KEAIESERGAIMLAVVGGKVSEGINFSDSMCRCVVMVGLPYPSPSDIELLERIKHIEG-- 752
Query 103 TNPKATADEAEEAKPAPSD--WPADAKQ----LQQQQREQHFVDYGLLQCMITVNQTIGR 156
+A +D + + D + D + L+ +R +Y CM VNQ+IGR
Sbjct 753 ---RADSDSIKPSVTLVDDSYYSGDVQAGFGVLKSCKRRGK--EYYENLCMKAVNQSIGR 807
Query 157 AIRHSKDYAAILLVD 171
AIRH KDYA+ILLVD
Sbjct 808 AIRHEKDYASILLVD 822
> tpv:TP03_0644 DNA helicase; K11273 chromosome transmission fidelity
protein 1 [EC:3.6.4.13]
Length=740
Score = 77.8 bits (190), Expect = 2e-14, Method: Composition-based stats.
Identities = 47/122 (38%), Positives = 71/122 (58%), Gaps = 9/122 (7%)
Query 50 KTAFLFCVMGGRLSEGVNFSNALARLLVVVGMPFPSVKDKIFILHKEHYNAIMTNPKATA 109
K A LF V GG SEGV+FS+ LARL+++VG+P+P K+ L +E+YN K+TA
Sbjct 586 KGAILFAVFGGSQSEGVDFSDDLARLVLLVGLPYPPDNIKL-KLKREYYN---KKCKSTA 641
Query 110 DEAEEAKPAPSDWPADAKQLQQQQREQHFVDYGLLQCMITVNQTIGRAIRHSKDYAAILL 169
+ + + + ++ + REQ L C TVNQ IGRA+RH D++ ++L
Sbjct 642 NTHTITITSEAGEQSVSENYSKLAREQR-----TLLCYKTVNQCIGRAMRHRDDFSGVVL 696
Query 170 VD 171
+D
Sbjct 697 LD 698
> hsa:1663 DDX11, CHL1, CHLR1, KRG2, MGC133249, MGC9335; DEAD/H
(Asp-Glu-Ala-Asp/His) box polypeptide 11 (EC:3.6.4.13); K11273
chromosome transmission fidelity protein 1 [EC:3.6.4.13]
Length=856
Score = 75.5 bits (184), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 44/124 (35%), Positives = 65/124 (52%), Gaps = 26/124 (20%)
Query 48 QSKTAFLFCVMGGRLSEGVNFSNALARLLVVVGMPFPSVKDKIFILHKEHYNAIMTNPKA 107
Q A L V+GG++SEG+NFS+ L R +V+VGMPFP+++ +A + A
Sbjct 715 QVTGALLLSVVGGKMSEGINFSDNLGRCVVMVGMPFPNIR-----------SAELQEKMA 763
Query 108 TADEAEEAKPAPSDWPADAKQLQQQQREQHFVDYGLLQCMITVNQTIGRAIRHSKDYAAI 167
D+ AP P ++ CM VNQ+IGRAIRH KD+A++
Sbjct 764 YLDQT--LPRAPGQAPPGKALVEN-------------LCMKAVNQSIGRAIRHQKDFASV 808
Query 168 LLVD 171
+L+D
Sbjct 809 VLLD 812
> cel:M03C11.2 hypothetical protein; K11273 chromosome transmission
fidelity protein 1 [EC:3.6.4.13]
Length=848
Score = 74.7 bits (182), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 42/123 (34%), Positives = 59/123 (47%), Gaps = 32/123 (26%)
Query 49 SKTAFLFCVMGGRLSEGVNFSNALARLLVVVGMPFPSVKDKIFILHKEHYNAIMTNPKAT 108
SK A LF V+GG++SEG+NF + L R ++V+G+P+P+ K +
Sbjct 715 SKGAILFAVVGGKMSEGINFCDELGRAVIVIGLPYPN--------------------KTS 754
Query 109 ADEAEEAKPAPSDWPADAKQLQQQQREQHFVDYGLLQCMITVNQTIGRAIRHSKDYAAIL 168
+ E K + P L + CM VNQ IGRAIRH +DYAA+
Sbjct 755 VELRERMKFLDTQMPNGGNLLYES------------LCMHAVNQAIGRAIRHRRDYAAVY 802
Query 169 LVD 171
L D
Sbjct 803 LFD 805
> cpv:cgd7_3510 helicase ; K11273 chromosome transmission fidelity
protein 1 [EC:3.6.4.13]
Length=775
Score = 73.9 bits (180), Expect = 2e-13, Method: Composition-based stats.
Identities = 47/120 (39%), Positives = 61/120 (50%), Gaps = 30/120 (25%)
Query 52 AFLFCVMGGRLSEGVNFSNALARLLVVVGMPFPSVKDKIFILHKEHYNAIMTNPKATADE 111
A LF V+ G LSEGVNFS+ L R L++V +PFP + +F KE Y + + D
Sbjct 640 AILFAVLNGTLSEGVNFSDELCRCLIIVSLPFPQKTEMLF--SKERYFN-----RGSCD- 691
Query 112 AEEAKPAPSDWPADAKQLQQQQREQHFVDYGLLQCMITVNQTIGRAIRHSKDYAAILLVD 171
R ++ Y + CM TVNQ IGRAIRH DY++ILLVD
Sbjct 692 --------------------YNRYENI--YRKVLCMKTVNQCIGRAIRHRNDYSSILLVD 729
> dre:492353 zgc:92172 (EC:3.6.4.13); K11273 chromosome transmission
fidelity protein 1 [EC:3.6.4.13]
Length=890
Score = 73.9 bits (180), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 44/125 (35%), Positives = 61/125 (48%), Gaps = 37/125 (29%)
Query 52 AFLFCVMGGRLSEGVNFSNALARLLVVVGMPFPSVK-----DKIFILHKEHYNAIMTNPK 106
A LF V+GG++SEG+NFS+ L R +V+VGMP+P++K +K+ L K + +P
Sbjct 752 ALLFSVVGGKMSEGINFSDDLGRCIVMVGMPYPNIKSPELQEKMAYLDKHMPHVAGKSPG 811
Query 107 ATADEAEEAKPAPSDWPADAKQLQQQQREQHFVDYGLLQCMITVNQTIGRAIRHSKDYAA 166
E+ CM VNQ+IGRAIRH DYA
Sbjct 812 KALVES--------------------------------LCMKAVNQSIGRAIRHRGDYAC 839
Query 167 ILLVD 171
I+L D
Sbjct 840 IVLCD 844
> mmu:320209 Ddx11, 4732462I11Rik, CHL1, CHLR1, KRG2, MGC90809;
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 11 (CHL1-like
helicase homolog, S. cerevisiae) (EC:3.6.4.13); K11273 chromosome
transmission fidelity protein 1 [EC:3.6.4.13]
Length=880
Score = 73.6 bits (179), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 43/132 (32%), Positives = 66/132 (50%), Gaps = 26/132 (19%)
Query 40 LSAQESSPQSKTAFLFCVMGGRLSEGVNFSNALARLLVVVGMPFPSVKDKIFILHKEHYN 99
+S S A L V+GG++SEG+NFS+ L R +V+VGMP+P++K + N
Sbjct 729 MSCSHSEGHLTGALLLSVVGGKMSEGINFSDDLGRCVVMVGMPYPNIKSPELQEKMAYLN 788
Query 100 AIMTNPKATADEAEEAKPAPSDWPADAKQLQQQQREQHFVDYGLLQCMITVNQTIGRAIR 159
T P+ + +P P + CM +NQ+IGRAIR
Sbjct 789 --QTLPRT------QGQPLPGTVLIENL------------------CMKAINQSIGRAIR 822
Query 160 HSKDYAAILLVD 171
H +D+A+I+L+D
Sbjct 823 HQRDFASIVLLD 834
> bbo:BBOV_I004620 19.m02044; DNA repair helicase (rad3) and DEAD_2
domain containing protein; K11273 chromosome transmission
fidelity protein 1 [EC:3.6.4.13]
Length=775
Score = 68.6 bits (166), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 42/142 (29%), Positives = 66/142 (46%), Gaps = 20/142 (14%)
Query 30 FQWEDKTCKALSAQESSPQSKTAFLFCVMGGRLSEGVNFSNALARLLVVVGMPFPSVKDK 89
F+ DK + A LF V GG SEGV+F + LARL+++VG+P+P K
Sbjct 603 FESRDKNVDTFGEYSKVALTTGAILFGVYGGSQSEGVDFHDGLARLVLLVGLPYPPETVK 662
Query 90 IFILHKEHYNAIMTNPKATADEAEEAKPAPSDWPADAKQLQQQQREQHFVDYGLLQCMIT 149
+ L + + + + + D P ++ D + C
Sbjct 663 L-RLRRSYL---------------KCRASQVDLPLKRREFYNMLSN----DISTITCYKI 702
Query 150 VNQTIGRAIRHSKDYAAILLVD 171
VNQ+IGRA+RH D+AA++L+D
Sbjct 703 VNQSIGRAMRHKDDFAALVLLD 724
> pfa:MAL13P1.134 DEAD box helicase, putative; K11273 chromosome
transmission fidelity protein 1 [EC:3.6.4.13]
Length=1099
Score = 64.7 bits (156), Expect = 1e-10, Method: Composition-based stats.
Identities = 41/121 (33%), Positives = 64/121 (52%), Gaps = 15/121 (12%)
Query 54 LFCVMGGRLSEGVNFSNALARLLVVVGMPF---PSVKDKIFILHKEHYNAIMTNPKATAD 110
LFCVM G+LSEG+NF + L R +VVVG+PF SV DK +L ++Y + T D
Sbjct 897 LFCVMNGKLSEGINFYDDLCRNVVVVGIPFIKHDSVVDK-NLLRLKYYREYTKDIMNTHD 955
Query 111 EAEEAKPAPSDWPADAKQLQQQQREQHFVDYGLLQCMITVNQTIGRAIRHSKDYAAILLV 170
+ + K D D ++ + ++ M +NQ IGR++RH D+++ +
Sbjct 956 KNDNTKILIYD---DVHKMCESYETKY--------AMKIINQCIGRSLRHVNDFSSFFFL 1004
Query 171 D 171
D
Sbjct 1005 D 1005
> cpv:cgd6_2660 DNA repair helicase ; K11136 regulator of telomere
elongation helicase 1
Length=1108
Score = 63.5 bits (153), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 39/123 (31%), Positives = 57/123 (46%), Gaps = 33/123 (26%)
Query 49 SKTAFLFCVMGGRLSEGVNFSNALARLLVVVGMPFPSVKDKIFILHKEHYNAIMTNPKAT 108
S + L V G++SEG+NFS+ R +++ G+PFPS+ D L K++
Sbjct 730 SSGSLLIAVCRGKVSEGINFSDNACRGVIIAGLPFPSIADARVCLKKQY----------- 778
Query 109 ADEAEEAKPAPSDWPADAKQLQQQQREQHFVDYGLLQCMITVNQTIGRAIRHSKDYAAIL 168
DE++ D +Q Q Q + VNQ IGR +RH DY AI+
Sbjct 779 MDESK----------MDGRQWYNQ------------QAIRAVNQAIGRVVRHRNDYGAII 816
Query 169 LVD 171
L D
Sbjct 817 LAD 819
> pfa:PF14_0081 DNA-repair helicase, putative; K11136 regulator
of telomere elongation helicase 1
Length=1160
Score = 60.5 bits (145), Expect = 2e-09, Method: Composition-based stats.
Identities = 36/124 (29%), Positives = 62/124 (50%), Gaps = 16/124 (12%)
Query 48 QSKTAFLFCVMGGRLSEGVNFSNALARLLVVVGMPFPSVKDKIFILHKEHYNAIMTNPKA 107
+ K A L V G++SEG++F + R +++ G+P+ +V D I KE + N K
Sbjct 700 KRKGAILMGVCRGKISEGIDFKDDCCRAVIICGLPYGNVYDSKIIFKKE----FLDNFKY 755
Query 108 TADEAEEAKPAPSDWPADAKQLQQQQREQHFVDYGLLQCMITVNQTIGRAIRHSKDYAAI 167
+ + +P S+ ++ + + E + M ++NQ+IGR IRH DY AI
Sbjct 756 ---QKTDERPNSSNITSNISKGNKWYNE---------EAMRSINQSIGRVIRHKNDYGAI 803
Query 168 LLVD 171
+D
Sbjct 804 FFLD 807
> mmu:237911 Brip1, 3110009N10Rik, 8030460J03Rik, Bach1, FACJ,
Fancj, OF; BRCA1 interacting protein C-terminal helicase 1
(EC:3.6.4.13)
Length=1174
Score = 58.5 bits (140), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 40/128 (31%), Positives = 57/128 (44%), Gaps = 44/128 (34%)
Query 52 AFLFCVMGGRLSEGVNFSNALARLLVVVGMPFPSVKDKIFILHKEHYNAIMTNPKATADE 111
A L V G++SEG++FS+ AR ++ VG+PFP+VKD L E
Sbjct 758 ALLIAVCRGKVSEGLDFSDDNARAVITVGIPFPNVKD----LQVE--------------- 798
Query 112 AEEAKPAPSDWPADAKQLQQQQREQHFVDYGLL--------QCMITVNQTIGRAIRHSKD 163
L++Q + H GLL Q +NQ +GR IRH D
Sbjct 799 -----------------LKRQYNDHHSKSRGLLPGRQWYEIQAYRALNQALGRCIRHKND 841
Query 164 YAAILLVD 171
+ A++LVD
Sbjct 842 WGALILVD 849
> tpv:TP04_0785 DNA repair helicase; K11136 regulator of telomere
elongation helicase 1
Length=962
Score = 58.5 bits (140), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 34/124 (27%), Positives = 57/124 (45%), Gaps = 26/124 (20%)
Query 48 QSKTAFLFCVMGGRLSEGVNFSNALARLLVVVGMPFPSVKDKIFILHKEHYNAIMTNPKA 107
Q + F + GRL+EG++FS+ R + V G+P+PS D L ++ + +
Sbjct 705 QGNGSVFFAICRGRLAEGIDFSDDYCRGIFVCGIPYPSRFDDNTALKMDYLDKL------ 758
Query 108 TADEAEEAKPAPSDWPADAKQLQQQQREQHFVDYGLLQCMITVNQTIGRAIRHSKDYAAI 167
+++ E ++W Q + +NQ +GR+IRH +DY AI
Sbjct 759 -SNKTYEKSNLANEWYTS-------------------QAIRAINQAVGRSIRHDRDYGAI 798
Query 168 LLVD 171
LL D
Sbjct 799 LLAD 802
> hsa:51750 RTEL1, C20orf41, DKFZp434C013, KIAA1088, NHL, RTEL,
bK3184A7.3; regulator of telomere elongation helicase 1 (EC:3.6.4.12);
K11136 regulator of telomere elongation helicase
1
Length=1219
Score = 57.4 bits (137), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 44/152 (28%), Positives = 65/152 (42%), Gaps = 27/152 (17%)
Query 20 LACNNRSKRGFQWEDKTCKALSAQESSPQSKTAFLFCVMGGRLSEGVNFSNALARLLVVV 79
L RSK F +T A A+ ++P S A V G+ SEG++FS+ R ++V
Sbjct 588 LFVEPRSKGSF---SETISAYYARVAAPGSTGATFLAVCRGKASEGLDFSDTNGRGVIVT 644
Query 80 GMPFPSVKDKIFILHKEHYNAIMTNPKATADEAEEAKPAPSDWPADAKQLQQQQREQHFV 139
G+P+P D +L + DE + A + + + +QQ
Sbjct 645 GLPYPPRMDPRVVLKMQFL-----------DEMKGQGGAGGQFLSGQEWYRQQASR---- 689
Query 140 DYGLLQCMITVNQTIGRAIRHSKDYAAILLVD 171
VNQ IGR IRH +DY A+ L D
Sbjct 690 ---------AVNQAIGRVIRHRQDYGAVFLCD 712
> dre:503732 rtel1; zgc:113114 (EC:3.6.4.12); K11136 regulator
of telomere elongation helicase 1
Length=1177
Score = 57.4 bits (137), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 37/125 (29%), Positives = 54/125 (43%), Gaps = 24/125 (19%)
Query 47 PQSKTAFLFCVMGGRLSEGVNFSNALARLLVVVGMPFPSVKDKIFILHKEHYNAIMTNPK 106
P S F V G+ SEG++F++ R +V+ G+PFP D +L ++ + + N
Sbjct 611 PNSSGGSFFAVCRGKASEGLDFADTYGRGVVITGLPFPPRMDPRVVLKMQYLDEMCRNKI 670
Query 107 ATADEAEEAKPAPSDWPADAKQLQQQQREQHFVDYGLLQCMITVNQTIGRAIRHSKDYAA 166
+ K L Q+ + Q VNQ IGR IRH +DY A
Sbjct 671 -----------------SGVKYLTGQEWYRQ-------QASRAVNQAIGRVIRHREDYGA 706
Query 167 ILLVD 171
I L D
Sbjct 707 IFLCD 711
> hsa:83990 BRIP1, BACH1, FANCJ, FLJ90232, MGC126521, MGC126523,
OF; BRCA1 interacting protein C-terminal helicase 1 (EC:3.6.4.13)
Length=1249
Score = 56.6 bits (135), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 39/128 (30%), Positives = 57/128 (44%), Gaps = 44/128 (34%)
Query 52 AFLFCVMGGRLSEGVNFSNALARLLVVVGMPFPSVKDKIFILHKEHYNAIMTNPKATADE 111
A L V G++SEG++FS+ AR ++ +G+PFP+VKD L E
Sbjct 755 ALLVAVCRGKVSEGLDFSDDNARAVITIGIPFPNVKD----LQVE--------------- 795
Query 112 AEEAKPAPSDWPADAKQLQQQQREQHFVDYGLL--------QCMITVNQTIGRAIRHSKD 163
L++Q + H GLL Q +NQ +GR IRH D
Sbjct 796 -----------------LKRQYNDHHSKLRGLLPGRQWYEIQAYRALNQALGRCIRHRND 838
Query 164 YAAILLVD 171
+ A++LVD
Sbjct 839 WGALILVD 846
> ath:AT1G20720 ATP binding / ATP-dependent DNA helicase/ ATP-dependent
helicase/ DNA binding / hydrolase, acting on acid
anhydrides, in phosphorus-containing anhydrides / nucleic acid
binding
Length=1175
Score = 56.2 bits (134), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 38/122 (31%), Positives = 58/122 (47%), Gaps = 28/122 (22%)
Query 50 KTAFLFCVMGGRLSEGVNFSNALARLLVVVGMPFPSVKDKIFILHKEHYNAIMTNPKATA 109
K A V G++SEG++F++ AR +++VG+PFP++ D I + K+ YN + K+
Sbjct 651 KGAAFLAVCRGKVSEGIDFADDNARAVIIVGIPFPNLHD-IQVGLKKKYNDTYKSSKSLL 709
Query 110 DEAEEAKPAPSDWPADAKQLQQQQREQHFVDYGLLQCMITVNQTIGRAIRHSKDYAAILL 169
S+W QQ R +NQ GR IRH DY AI+
Sbjct 710 --------GGSEW-----YCQQAYR--------------ALNQAAGRCIRHRFDYGAIIF 742
Query 170 VD 171
+D
Sbjct 743 LD 744
> dre:794038 brip1, fancj, si:ch211-158l18.1; BRCA1 interacting
protein C-terminal helicase 1
Length=1217
Score = 55.8 bits (133), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 37/120 (30%), Positives = 59/120 (49%), Gaps = 28/120 (23%)
Query 52 AFLFCVMGGRLSEGVNFSNALARLLVVVGMPFPSVKDKIFILHKEHYNAIMTNPKATADE 111
A L V G++SEG++F++ AR +V +G+PFP++KD + + K YN
Sbjct 788 ALLVAVCRGKVSEGLDFTDDNARAVVTIGIPFPNIKD-LQVELKMKYN------------ 834
Query 112 AEEAKPAPSDWPADAKQLQQQQREQHFVDYGLLQCMITVNQTIGRAIRHSKDYAAILLVD 171
D A ++ L R + +Q +NQ +GR IRH D+ A++LVD
Sbjct 835 ---------DKHAKSRGLLPGGR------WYEIQAYRALNQALGRCIRHRNDWGALILVD 879
> mmu:269400 Rtel1, AI451565, AW540478, Rtel; regulator of telomere
elongation helicase 1 (EC:3.6.4.12); K11136 regulator
of telomere elongation helicase 1
Length=1209
Score = 53.9 bits (128), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 45/152 (29%), Positives = 62/152 (40%), Gaps = 27/152 (17%)
Query 20 LACNNRSKRGFQWEDKTCKALSAQESSPQSKTAFLFCVMGGRLSEGVNFSNALARLLVVV 79
L R+K F + A Q +SP S A V G+ SEG++FS+ R ++V
Sbjct 588 LFVEPRNKGSF---SEVIDAYYQQVASPASNGATFLAVCRGKASEGLDFSDMNGRGVIVT 644
Query 80 GMPFPSVKDKIFILHKEHYNAIMTNPKATADEAEEAKPAPSDWPADAKQLQQQQREQHFV 139
G+P+P D +L K + M + + +W QQQ +
Sbjct 645 GLPYPPRMDPRVVL-KMQFLDEMRGRSGVGGQCLSGQ----EW-------YQQQASR--- 689
Query 140 DYGLLQCMITVNQTIGRAIRHSKDYAAILLVD 171
VNQ IGR IRH DY AI L D
Sbjct 690 ---------AVNQAIGRVIRHRHDYGAIFLCD 712
> cel:F33H2.1 dog-1; Deletions Of G-rich DNA family member (dog-1)
Length=983
Score = 51.6 bits (122), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 32/122 (26%), Positives = 57/122 (46%), Gaps = 32/122 (26%)
Query 52 AFLFCVMGGRLSEGVNFSNALARLLVVVGMPFPSVKDKIFILHKEHYNAIMTNPKA--TA 109
+ +F V G++SEG++F++ AR+++ VG+P+P+ D + K+ YN + K T
Sbjct 832 SLMFAVFRGKVSEGIDFADDRARVVISVGIPYPNAMDDQ-VNAKKLYNDQNSKEKGILTG 890
Query 110 DEAEEAKPAPSDWPADAKQLQQQQREQHFVDYGLLQCMITVNQTIGRAIRHSKDYAAILL 169
DE + Q +NQ +GR +RH D+ A+L+
Sbjct 891 DE-----------------------------WYTTQAYRALNQALGRCLRHKNDWGAMLM 921
Query 170 VD 171
+D
Sbjct 922 ID 923
> ath:AT1G20750 helicase-related
Length=1179
Score = 50.4 bits (119), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 40/129 (31%), Positives = 63/129 (48%), Gaps = 30/129 (23%)
Query 42 AQESSPQSKTAFLFCVMGGRLSEGVNFSNALARLLVVVGMPFPSVKDKIFILHKEHYNAI 101
AQ+ S + +AFL V G++SEG++FS+ AR +++VG+P P++ D I + K YN
Sbjct 616 AQDDSKRG-SAFL-AVCRGKVSEGMDFSDDNARAVIIVGIPLPNLGD-ILVELKRKYN-- 670
Query 102 MTNPKATADEAEEAKPAPSDWPADAKQLQQQQREQHFVDYGLLQCMITVNQTIGRAIRHS 161
+++ + S+W QQ R +NQ GR IRH
Sbjct 671 ------DTNKSSKNLLGGSEW-----YCQQAYR--------------ALNQAAGRCIRHR 705
Query 162 KDYAAILLV 170
DY AI+ +
Sbjct 706 FDYGAIIFL 714
> ath:AT1G79950 helicase-related; K11136 regulator of telomere
elongation helicase 1
Length=1040
Score = 49.7 bits (117), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 33/118 (27%), Positives = 50/118 (42%), Gaps = 21/118 (17%)
Query 54 LFCVMGGRLSEGVNFSNALARLLVVVGMPFPSVKDKIFILHKEHYNAIMTNPKATADEAE 113
F V G++SEG++F++ R +V+ G+P+ V D L +E + E
Sbjct 635 FFAVCRGKVSEGLDFADGAGRAVVITGLPYARVTDPRVKLKREFLD-------------E 681
Query 114 EAKPAPSDWPADAKQLQQQQREQHFVDYGLLQCMITVNQTIGRAIRHSKDYAAILLVD 171
+++ A P Q + VNQ IGR IRH DY AI+ D
Sbjct 682 QSQLADVKLPRSTLLSGSMWYSQ--------EAARAVNQAIGRVIRHRHDYGAIIFCD 731
> tgo:TGME49_009770 DNA repair helicase, putative ; K11136 regulator
of telomere elongation helicase 1
Length=1649
Score = 45.8 bits (107), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 31/125 (24%), Positives = 55/125 (44%), Gaps = 17/125 (13%)
Query 52 AFLFCVMGGRLSEGVNFSNALARLLVVVGMPFPSVKDKIFILHKEHYNAIMTNPKAT--- 108
A L V G+ +EG++FS+ R +V+ G+P S + L + + M A
Sbjct 965 AVLLAVCKGKAAEGIDFSDHACRAVVICGLPLASFFEPRVQLKRMWLDDCMQKLVAETGG 1024
Query 109 ADEAEEAKPAPSDWPA--DAKQLQQQQREQHFVDYGLLQCMITVNQTIGRAIRHSKDYAA 166
A +E + P + +Q Q+ + VNQ +GR +RH+KD+
Sbjct 1025 ASRGKEQGTGETTAPVLINGRQWYDQEARR------------AVNQAVGRVVRHAKDFGV 1072
Query 167 ILLVD 171
++ +D
Sbjct 1073 VVFLD 1077
> bbo:BBOV_III008820 17.m07769; DNA repair helicase (rad3) family
protein; K11136 regulator of telomere elongation helicase
1
Length=948
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 33/112 (29%), Positives = 50/112 (44%), Gaps = 29/112 (25%)
Query 60 GRLSEGVNFSNALARLLVVVGMPFPSVKDKIFILHKEHYNAIMTNPKATADEAEEAKPAP 119
G+L+EG++FS+ R + + G+P+P NP EE
Sbjct 706 GKLAEGIDFSDDSCRGVFLCGVPYP-------------------NP------YEETIALK 740
Query 120 SDWPADAKQLQQQQREQHFVDYGLLQCMITVNQTIGRAIRHSKDYAAILLVD 171
D+ ++L + E + + Q + VNQ IGR IRH DY AI+L D
Sbjct 741 MDY---LRKLYGKNNE-NVTHWFTSQAIRAVNQAIGRVIRHINDYGAIVLAD 788
> cel:F25H2.13 rtel-1; RTEL (mammalian Regulator of TELomere length)
homolog family member (rtel-1); K11136 regulator of telomere
elongation helicase 1
Length=994
Score = 42.7 bits (99), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 32/120 (26%), Positives = 60/120 (50%), Gaps = 24/120 (20%)
Query 52 AFLFCVMGGRLSEGVNFSNALARLLVVVGMPFPSVKDKIFILHKEHYNAIMTNPKATADE 111
A L V G++SEG++F +A +R ++++G+P+P + D+ +L K + + +M
Sbjct 634 AALLAVCRGKVSEGIDFCDAESRAVIIIGIPYPPIHDERVVLKKMYLDDLMGR------- 686
Query 112 AEEAKPAPSDWPADAKQLQQQQREQHFVDYGLLQCMITVNQTIGRAIRHSKDYAAILLVD 171
D K +Q + D+ ++ VNQ IGR +RH D+ ++L+D
Sbjct 687 ------------KDTKSERQSSQ-----DWYQMEAFRAVNQAIGRVLRHKDDFGTVVLMD 729
> tgo:TGME49_031800 hypothetical protein
Length=2272
Score = 42.0 bits (97), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 26/87 (29%), Positives = 47/87 (54%), Gaps = 14/87 (16%)
Query 44 ESSPQSKTAFLFCVMGGRLSEGVNFSNALARLLVVVGMPFPSVKDKIFILHKEHYN---- 99
ES + A + V G++SEG++F++A R ++ +G+P+P+++D I K +N
Sbjct 1677 ESVDERGHALMLAVYRGKMSEGLSFNDAYCRGVLCIGIPYPALRDPK-IESKMKFNDALH 1735
Query 100 AIMTNPK---------ATADEAEEAKP 117
A+ +P+ ATAD A+P
Sbjct 1736 ALSLSPEHLLSCASDMATADRTTSARP 1762
Score = 36.6 bits (83), Expect = 0.037, Method: Compositional matrix adjust.
Identities = 13/29 (44%), Positives = 19/29 (65%), Gaps = 0/29 (0%)
Query 143 LLQCMITVNQTIGRAIRHSKDYAAILLVD 171
++Q +NQ IGR IRH DY ++L+D
Sbjct 1858 IIQAYRALNQAIGRCIRHKHDYGVVILLD 1886
> hsa:2068 ERCC2, COFS2, EM9, MGC102762, MGC126218, MGC126219,
TTD, XPD; excision repair cross-complementing rodent repair
deficiency, complementation group 2 (EC:3.6.4.12); K10844 DNA
excision repair protein ERCC-2 [EC:3.6.4.12]
Length=760
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 30/129 (23%), Positives = 50/129 (38%), Gaps = 32/129 (24%)
Query 43 QESSPQSKTAFLFCVMGGRLSEGVNFSNALARLLVVVGMPFPSVKDKIFILHKEHYNAIM 102
QE+ + A L V G++SEG++F + R +++ G+P+ + +I E+
Sbjct 585 QEACENGRGAILLSVARGKVSEGIDFVHHYGRAVIMFGVPYVYTQSRILKARLEYLRD-- 642
Query 103 TNPKATADEAEEAKPAPSDWPADAKQLQQQQREQHFVDYGLLQCMITVNQTIGRAIRHSK 162
Q Q RE F+ + M Q +GRAIR
Sbjct 643 ---------------------------QFQIRENDFLTF---DAMRHAAQCVGRAIRGKT 672
Query 163 DYAAILLVD 171
DY ++ D
Sbjct 673 DYGLMVFAD 681
> mmu:13871 Ercc2, AA407812, AU020867, AW240756, Ercc-2, XPD;
excision repair cross-complementing rodent repair deficiency,
complementation group 2 (EC:3.6.4.12); K10844 DNA excision
repair protein ERCC-2 [EC:3.6.4.12]
Length=760
Score = 40.4 bits (93), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 30/129 (23%), Positives = 50/129 (38%), Gaps = 32/129 (24%)
Query 43 QESSPQSKTAFLFCVMGGRLSEGVNFSNALARLLVVVGMPFPSVKDKIFILHKEHYNAIM 102
QE+ + A L V G++SEG++F + R +++ G+P+ + +I E+
Sbjct 585 QEACENGRGAILLSVARGKVSEGIDFVHHYGRAVIMFGVPYVYTQSRILKARLEYLRD-- 642
Query 103 TNPKATADEAEEAKPAPSDWPADAKQLQQQQREQHFVDYGLLQCMITVNQTIGRAIRHSK 162
Q Q RE F+ + M Q +GRAIR
Sbjct 643 ---------------------------QFQIRENDFLTF---DAMRHAAQCVGRAIRGKT 672
Query 163 DYAAILLVD 171
DY ++ D
Sbjct 673 DYGLMVFAD 681
> cel:Y50D7A.2 hypothetical protein; K10844 DNA excision repair
protein ERCC-2 [EC:3.6.4.12]
Length=606
Score = 39.7 bits (91), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 15/48 (31%), Positives = 31/48 (64%), Gaps = 0/48 (0%)
Query 44 ESSPQSKTAFLFCVMGGRLSEGVNFSNALARLLVVVGMPFPSVKDKIF 91
E+ + A LF V G++SEG++FS+ L R ++++G+P+ + ++
Sbjct 431 EACDSGRGAVLFSVARGKVSEGIDFSHHLGRCVIMLGIPYMYTESRVL 478
> ath:AT1G03190 UVH6; UVH6 (ULTRAVIOLET HYPERSENSITIVE 6); ATP
binding / ATP-dependent DNA helicase/ ATP-dependent helicase/
DNA binding / hydrolase, acting on acid anhydrides, in phosphorus-containing
anhydrides / nucleic acid binding; K10844
DNA excision repair protein ERCC-2 [EC:3.6.4.12]
Length=758
Score = 38.1 bits (87), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 28/122 (22%), Positives = 46/122 (37%), Gaps = 32/122 (26%)
Query 50 KTAFLFCVMGGRLSEGVNFSNALARLLVVVGMPFPSVKDKIFILHKEHYNAIMTNPKATA 109
+ A F V G+++EG++F RL+V+ G+PF KI E+ +
Sbjct 592 RGAVFFSVARGKVAEGIDFDRHYGRLVVMYGVPFQYTLSKILRARLEYLHDTF------- 644
Query 110 DEAEEAKPAPSDWPADAKQLQQQQREQHFVDYGLLQCMITVNQTIGRAIRHSKDYAAILL 169
Q +E F+ + L+ Q +GR IR DY ++
Sbjct 645 ----------------------QIKEGDFLTFDALR---QAAQCVGRVIRSKADYGMMIF 679
Query 170 VD 171
D
Sbjct 680 AD 681
> sce:YER171W RAD3, REM1; 5' to 3' DNA helicase, involved in nucleotide
excision repair and transcription; subunit of RNA
polymerase II transcription initiation factor TFIIH; subunit
of Nucleotide Excision Repair Factor 3 (NEF3); homolog of human
XPD protein (EC:3.6.1.-); K10844 DNA excision repair protein
ERCC-2 [EC:3.6.4.12]
Length=778
Score = 37.4 bits (85), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 28/122 (22%), Positives = 49/122 (40%), Gaps = 32/122 (26%)
Query 50 KTAFLFCVMGGRLSEGVNFSNALARLLVVVGMPFPSVKDKIFILHKEHYNAIMTNPKATA 109
+ A L V G++SEG++F + R ++++G+PF + +I KA
Sbjct 594 RGAILLSVARGKVSEGIDFDHQYGRTVLMIGIPFQYTESRIL--------------KARL 639
Query 110 DEAEEAKPAPSDWPADAKQLQQQQREQHFVDYGLLQCMITVNQTIGRAIRHSKDYAAILL 169
+ E + RE F+ + M Q +GR +R DY ++L
Sbjct 640 EFMRE---------------NYRIRENDFLSF---DAMRHAAQCLGRVLRGKDDYGVMVL 681
Query 170 VD 171
D
Sbjct 682 AD 683
> cpv:cgd7_820 RAD3'DEXDc+HELICc protein' ; K10844 DNA excision
repair protein ERCC-2 [EC:3.6.4.12]
Length=841
Score = 36.6 bits (83), Expect = 0.047, Method: Compositional matrix adjust.
Identities = 29/126 (23%), Positives = 46/126 (36%), Gaps = 40/126 (31%)
Query 50 KTAFLFCVMGGRLSEGVNFSNALARLLVVVGMPFPSVKDKI----FILHKEHYNAIMTNP 105
+ A F + G+++EG++F R +V+VG+P+ KI KE+Y I N
Sbjct 694 RGAIFFSIARGKVAEGIDFDRHYGRCVVMVGIPYQYTLSKILQSRLSFLKENY-GIQENE 752
Query 106 KATADEAEEAKPAPSDWPADAKQLQQQQREQHFVDYGLLQCMITVNQTIGRAIRHSKDYA 165
T D +A +Q +GR IR DY
Sbjct 753 FLTFDAMRQA-----------------------------------SQCVGRVIRSKADYG 777
Query 166 AILLVD 171
++ D
Sbjct 778 LMIFAD 783
> tgo:TGME49_016870 excision repair protein rad15, putative ;
K10844 DNA excision repair protein ERCC-2 [EC:3.6.4.12]
Length=1065
Score = 35.8 bits (81), Expect = 0.069, Method: Compositional matrix adjust.
Identities = 25/122 (20%), Positives = 43/122 (35%), Gaps = 32/122 (26%)
Query 50 KTAFLFCVMGGRLSEGVNFSNALARLLVVVGMPFPSVKDKIFILHKEHYNAIMTNPKATA 109
+ A F + G+++EG++F R +V+ G+PF ++ KA
Sbjct 862 RGAVFFSIARGKVAEGIDFDRHFGRCVVLFGVPFQYTLSRVL--------------KARL 907
Query 110 DEAEEAKPAPSDWPADAKQLQQQQREQHFVDYGLLQCMITVNQTIGRAIRHSKDYAAILL 169
D E P + ++ M Q +GR IR DY ++
Sbjct 908 DFIREHYQIPDN------------------EFLTFDAMRQAAQCVGRVIRSKNDYGLMIF 949
Query 170 VD 171
D
Sbjct 950 AD 951
> pfa:PFI1650w DNA excision-repair helicase, putative; K10844
DNA excision repair protein ERCC-2 [EC:3.6.4.12]
Length=1056
Score = 33.9 bits (76), Expect = 0.29, Method: Composition-based stats.
Identities = 28/126 (22%), Positives = 46/126 (36%), Gaps = 40/126 (31%)
Query 50 KTAFLFCVMGGRLSEGVNFSNALARLLVVVGMPFPSVKDKIFI----LHKEHYNAIMTNP 105
K A + G+++EG++F + +++ G+P+ KI KE YN I N
Sbjct 887 KGAVFLSICRGKIAEGIDFDKHYGKCVILFGIPYQYTLSKILKSRLDFLKETYN-IQENE 945
Query 106 KATADEAEEAKPAPSDWPADAKQLQQQQREQHFVDYGLLQCMITVNQTIGRAIRHSKDYA 165
T D +A +Q +GR IR+ KDY
Sbjct 946 FLTFDAMRQA-----------------------------------SQCVGRIIRNKKDYG 970
Query 166 AILLVD 171
++ D
Sbjct 971 IMIFSD 976
> tpv:TP01_0155 DNA repair protein Rad3; K10844 DNA excision repair
protein ERCC-2 [EC:3.6.4.12]
Length=894
Score = 32.7 bits (73), Expect = 0.55, Method: Compositional matrix adjust.
Identities = 11/42 (26%), Positives = 25/42 (59%), Gaps = 0/42 (0%)
Query 50 KTAFLFCVMGGRLSEGVNFSNALARLLVVVGMPFPSVKDKIF 91
+ A + G+++EG++F++ R +++VG+PF K+
Sbjct 728 RGALFLSICRGKVAEGIDFNSHYGRCVILVGIPFQYTLSKVL 769
> hsa:100287029 DDX11L10, DDX11L1, DDX11P; DEAD/H (Asp-Glu-Ala-Asp/His)
box polypeptide 11 like 10
Length=133
Score = 32.3 bits (72), Expect = 0.72, Method: Compositional matrix adjust.
Identities = 15/34 (44%), Positives = 22/34 (64%), Gaps = 6/34 (17%)
Query 62 LSEGVNFSNALARLL------VVVGMPFPSVKDK 89
+SE +NFS+ L +LL V+ GMPFPS++
Sbjct 1 MSESINFSDNLGQLLSPPRCVVMPGMPFPSIRSP 34
> hsa:100507301 hypothetical protein LOC100507301
Length=133
Score = 32.3 bits (72), Expect = 0.75, Method: Compositional matrix adjust.
Identities = 15/34 (44%), Positives = 22/34 (64%), Gaps = 6/34 (17%)
Query 62 LSEGVNFSNALARLL------VVVGMPFPSVKDK 89
+SE +NFS+ L +LL V+ GMPFPS++
Sbjct 1 MSESINFSDNLGQLLSPPRCVVMPGMPFPSIRSP 34
> bbo:BBOV_III006210 17.m07551; hypothetical protein
Length=1062
Score = 31.2 bits (69), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 30/118 (25%), Positives = 45/118 (38%), Gaps = 37/118 (31%)
Query 54 LFCVMGGRLSEGVNFSNALARLLVVVGMPFPSVKDKIFILHKEHYNAIMTNPKATADEAE 113
+F V SEG+N L++VG+P+PS
Sbjct 945 MFAVCRSHSSEGLNLK---LSSLILVGLPYPSTI-------------------------- 975
Query 114 EAKPAPSDWPADAKQLQQQQREQHFVDYGLLQCMITVNQTIGRAIRHSKDYAAILLVD 171
AP D + + Q + ++ L + VNQ IGR IR+ KD I+L+D
Sbjct 976 ----APR---VDIARRYNRASGQKY-NWYLRETFRAVNQAIGRCIRNKKDRGVIILMD 1025
> hsa:100289775 hypothetical protein LOC100289775
Length=661
Score = 31.2 bits (69), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 13/43 (30%), Positives = 24/43 (55%), Gaps = 0/43 (0%)
Query 76 LVVVGMPFPSVKDKIFILHKEHYNAIMTNPKATADEAEEAKPA 118
L+ V MP P + +I L KEH + + +P+ + + + A+P
Sbjct 546 LMTVTMPLPLLGQQITSLQKEHLSLYVASPRLSHLQGQGAQPG 588
> bbo:BBOV_IV001300 21.m02831; DNA excision repair helicase; K10844
DNA excision repair protein ERCC-2 [EC:3.6.4.12]
Length=822
Score = 30.8 bits (68), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 9/34 (26%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 50 KTAFLFCVMGGRLSEGVNFSNALARLLVVVGMPF 83
+ A + G+++EG++F R ++++G+PF
Sbjct 654 RGALFLSICRGKVAEGIDFDRHYGRCVILIGVPF 687
> dre:415185 zgc:86905; K04441 p38 MAP kinase [EC:2.7.11.24]
Length=361
Score = 30.4 bits (67), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 17/48 (35%), Positives = 24/48 (50%), Gaps = 5/48 (10%)
Query 56 CVMGGRLS-----EGVNFSNALARLLVVVGMPFPSVKDKIFILHKEHY 98
C+MG L G ++ + L R++ VVG P P V KI H + Y
Sbjct 210 CIMGELLKGKVLFPGNDYIDQLKRIMEVVGTPTPDVLKKISSEHAQKY 257
> hsa:57645 POGK, BASS2, KIAA1513, KIAA15131, KRBOX2; pogo transposable
element with KRAB domain
Length=609
Score = 30.0 bits (66), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 14/37 (37%), Positives = 18/37 (48%), Gaps = 0/37 (0%)
Query 107 ATADEAEEAKPAPSDWPADAKQLQQQQREQHFVDYGL 143
T+ E EE+ P DWP Q + QHF +GL
Sbjct 116 GTSAENEESDVKPPDWPNPMNATSQFPQPQHFDSFGL 152
> tgo:TGME49_039000 hypothetical protein
Length=545
Score = 29.3 bits (64), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 17/58 (29%), Positives = 22/58 (37%), Gaps = 0/58 (0%)
Query 102 MTNPKATADEAEEAKPAPSDWPADAKQLQQQQREQHFVDYGLLQCMITVNQTIGRAIR 159
M K T DE E + W + + R + +GL QC I N T G R
Sbjct 1 MKEAKGTCDEGCEERDGQRGWESWGVPTDPESRANEQLGHGLEQCRIVSNLTSGEKAR 58
Lambda K H
0.319 0.131 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4276754328
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40