bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2212_orf1
Length=176
Score E
Sequences producing significant alignments: (Bits) Value
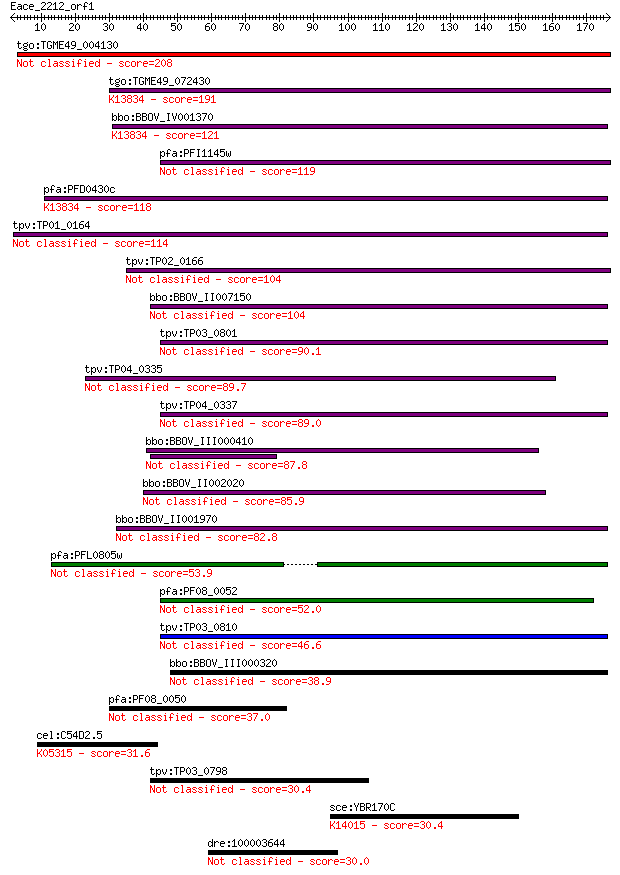
tgo:TGME49_004130 membrane-attack complex / perforin domain-co... 208 6e-54
tgo:TGME49_072430 membrane-attack complex / perforin domain-co... 191 9e-49
bbo:BBOV_IV001370 21.m02755; MAC/perforin domain containing pr... 121 1e-27
pfa:PFI1145w MAC/Perforin, putative 119 5e-27
pfa:PFD0430c MAC/Perforin, putative; K13834 sporozoite microne... 118 9e-27
tpv:TP01_0164 hypothetical protein 114 1e-25
tpv:TP02_0166 hypothetical protein 104 1e-22
bbo:BBOV_II007150 18.m06592; mac/perforin domain containing pr... 104 2e-22
tpv:TP03_0801 hypothetical protein 90.1 3e-18
tpv:TP04_0335 hypothetical protein 89.7 5e-18
tpv:TP04_0337 hypothetical protein 89.0 7e-18
bbo:BBOV_III000410 hypothetical protein 87.8 2e-17
bbo:BBOV_II002020 18.m06160; mac/perforin domain containing pr... 85.9 7e-17
bbo:BBOV_II001970 18.m09950; mac/perforin domain containing me... 82.8 5e-16
pfa:PFL0805w MAC/Perforin, putative 53.9 3e-07
pfa:PF08_0052 perforin like protein 5 52.0
tpv:TP03_0810 hypothetical protein 46.6 4e-05
bbo:BBOV_III000320 17.m10445; hypothetical protein 38.9 0.009
pfa:PF08_0050 MAC/Perforin, putative 37.0 0.036
cel:C54D2.5 cca-1; Calcium Channel, Alpha subunit family membe... 31.6 1.4
tpv:TP03_0798 hypothetical protein 30.4 3.2
sce:YBR170C NPL4, HRD4; Endoplasmic reticulum and nuclear memb... 30.4 3.5
dre:100003644 fc38h03; wu:fc38h03 30.0 3.8
> tgo:TGME49_004130 membrane-attack complex / perforin domain-containing
protein
Length=1054
Score = 208 bits (530), Expect = 6e-54, Method: Compositional matrix adjust.
Identities = 97/174 (55%), Positives = 126/174 (72%), Gaps = 2/174 (1%)
Query 3 QAAQDLEARREERRKQEWEAAQQAVTMQDIYARAKTVSPGFCYLGVGYDIVRGNPLGDPN 62
++ DL RE RR E A A + +Y +A P YLG GYD VRGNP+GDP+
Sbjct 332 ESVADLNRMRENRRIAEENRA--AAPLSAVYTKATKTVPAINYLGAGYDHVRGNPVGDPS 389
Query 63 LMGDPGLRSPVIRFSYSQDEEGVSSDLRELQPRGAYSRPFVACKQSENLSEVSSLADYQK 122
MGDPG+R PV+RF+Y+Q+E+GVS+DL LQP G Y R +VAC+QSE +SE+S+L+DYQ
Sbjct 390 SMGDPGIRPPVLRFTYAQNEDGVSNDLTVLQPLGGYVRQYVACRQSETISELSNLSDYQN 449
Query 123 ELEADASLVGGDSVGLNSFSASAAYRDIAKRTVKRNTKTFILKTYCLRFEAGLA 176
EL DASL GGD +GLNSFSAS YRD AK K++T+T++LK YC+R+EAG+A
Sbjct 450 ELSVDASLQGGDPIGLNSFSASTGYRDFAKEVSKKDTRTYMLKNYCMRYEAGVA 503
> tgo:TGME49_072430 membrane-attack complex / perforin domain-containing
protein ; K13834 sporozoite microneme protein 2
Length=854
Score = 191 bits (486), Expect = 9e-49, Method: Compositional matrix adjust.
Identities = 82/147 (55%), Positives = 115/147 (78%), Gaps = 0/147 (0%)
Query 30 QDIYARAKTVSPGFCYLGVGYDIVRGNPLGDPNLMGDPGLRSPVIRFSYSQDEEGVSSDL 89
+ +Y RA +P +LGVGYD ++GNP+GDP++M DPGLRSP+I FS+ QD +GV++DL
Sbjct 220 ESMYTRAVETAPATNFLGVGYDSIKGNPIGDPDMMVDPGLRSPIIVFSFQQDPDGVTNDL 279
Query 90 RELQPRGAYSRPFVACKQSENLSEVSSLADYQKELEADASLVGGDSVGLNSFSASAAYRD 149
LQP GA++RPF AC+QSEN++E+ +L+DYQK L DA+L GGDS+G+NSFS S Y++
Sbjct 280 NYLQPLGAFTRPFSACRQSENVNELDTLSDYQKVLSVDAALHGGDSLGINSFSGSTGYKE 339
Query 150 IAKRTVKRNTKTFILKTYCLRFEAGLA 176
A+ + K+F+LKTYC+R+EAGLA
Sbjct 340 FAQDVSSKANKSFMLKTYCIRYEAGLA 366
> bbo:BBOV_IV001370 21.m02755; MAC/perforin domain containing
protein; K13834 sporozoite microneme protein 2
Length=978
Score = 121 bits (303), Expect = 1e-27, Method: Composition-based stats.
Identities = 63/152 (41%), Positives = 97/152 (63%), Gaps = 12/152 (7%)
Query 31 DIYARAKTVSPGFC----YLGVGYDIVRGNPLGDPNLMGDPGLRSPVIRFSYSQ---DEE 83
D+ ++PG YLG GYDI+ GNPLGDP +M DPG R PV+R +S+ + +
Sbjct 312 DVTDAEGRLNPGLAAAMRYLGSGYDIIYGNPLGDPVIMVDPGYRHPVLRLDWSEKYYNND 371
Query 84 GVSSDLRELQPRGAYSRPFVACKQSENLSEVSSLADYQKELEADASLVGGDSVGLNSFSA 143
G ++++E P+G + RP ++C+QSE++ ++++ DY+KEL DA + D SFSA
Sbjct 372 G--ANMKE--PKGGWIRPELSCRQSESVDHINTMDDYKKELSVDAKM-SADMPLYFSFSA 426
Query 144 SAAYRDIAKRTVKRNTKTFILKTYCLRFEAGL 175
SA Y+++ + TK +ILKTYCLR+ AG+
Sbjct 427 SAGYKNMVRTLATNETKNYILKTYCLRYVAGI 458
> pfa:PFI1145w MAC/Perforin, putative
Length=821
Score = 119 bits (298), Expect = 5e-27, Method: Composition-based stats.
Identities = 55/133 (41%), Positives = 84/133 (63%), Gaps = 3/133 (2%)
Query 45 YLGVGYDIVRGNPLGDPNLMGDPGLRSPVIRFSYSQDEEGVSSDLRELQPRGAYSRPFVA 104
YLGVGYD + GNP+GDP L DPG R +I+ +Y + +E + + P G++ R ++
Sbjct 241 YLGVGYDFIFGNPIGDPFLKVDPGYRDSIIKLTYPKSDEDYPDNYMNINPNGSFVRNEIS 300
Query 105 CKQSENLSEVSSLADYQKELEADASLVGGDSVGL-NSFSASAAYRDIAKRTVKRNTKTFI 163
C +SE SE+S++++Y KEL DAS+ G S GL SFSAS Y+ ++ K + F+
Sbjct 301 CNRSEKESEISTMSEYTKELSVDASI--GASYGLFGSFSASTGYKSVSNTISKNKFRMFM 358
Query 164 LKTYCLRFEAGLA 176
LK+YC ++ A L+
Sbjct 359 LKSYCFKYVASLS 371
> pfa:PFD0430c MAC/Perforin, putative; K13834 sporozoite microneme
protein 2
Length=842
Score = 118 bits (296), Expect = 9e-27, Method: Composition-based stats.
Identities = 55/165 (33%), Positives = 101/165 (61%), Gaps = 1/165 (0%)
Query 11 RREERRKQEWEAAQQAVTMQDIYARAKTVSPGFCYLGVGYDIVRGNPLGDPNLMGDPGLR 70
+++E ++ + + + ++I ++ +V PG ++G+GY+++ GNPLG+ + + DPG R
Sbjct 212 QKDEDNEENDDKDENTLENRNIISKHTSVFPGLYFIGIGYNLLFGNPLGEADSLIDPGYR 271
Query 71 SPVIRFSYSQDEEGVSSDLRELQPRGAYSRPFVACKQSENLSEVSSLADYQKELEADASL 130
+ + ++ +EG+++DL LQP + R AC + E+++E SS++DY K L A+A
Sbjct 272 AQIYLMEWALSKEGIANDLSTLQPVNGWIRKENACSRVESITECSSISDYTKSLSAEAK- 330
Query 131 VGGDSVGLNSFSASAAYRDIAKRTVKRNTKTFILKTYCLRFEAGL 175
V G G+ SFSAS Y KR+ KTF++K+ C+++ GL
Sbjct 331 VSGSYWGIASFSASTGYSSFLHEVTKRSKKTFLVKSNCVKYTIGL 375
> tpv:TP01_0164 hypothetical protein
Length=1182
Score = 114 bits (286), Expect = 1e-25, Method: Composition-based stats.
Identities = 66/176 (37%), Positives = 100/176 (56%), Gaps = 19/176 (10%)
Query 2 AQAAQDLEARREERRKQ-EWEAAQQAVTMQDIYARAKTVSPGFCYLGVGYDIVRGNPLGD 60
+ A+ + R + R Q +W + A +M+ YLG GYDI+ GNPLGD
Sbjct 559 GETAKRVTTRIDPRDPQMDWNNSGLAASMR--------------YLGSGYDIIFGNPLGD 604
Query 61 PNLMGDPGLRSPVIRFSYSQDEEGVSSDLREL-QPRGAYSRPFVACKQSENLSEVSSLAD 119
P +M D G R+PVIR ++ ++E ++ D L +PRG++ RP +C+QSE + V+++ D
Sbjct 605 PVVMMDQGYRNPVIRLNW--EDEYLNKDGANLKEPRGSWIRPEYSCRQSETIDHVNTVDD 662
Query 120 YQKELEADASLVGGDSVGLNSFSASAAYRDIAKRTVKRNTKTFILKTYCLRFEAGL 175
++KEL DA G SFSAS Y++ K T +T+I KTYCLR+ G+
Sbjct 663 FKKELSVDAQASYGIPY-FFSFSASTGYKNFVKSTATNKVRTYITKTYCLRYVGGI 717
> tpv:TP02_0166 hypothetical protein
Length=812
Score = 104 bits (260), Expect = 1e-22, Method: Composition-based stats.
Identities = 57/142 (40%), Positives = 87/142 (61%), Gaps = 4/142 (2%)
Query 35 RAKTVSPGFCYLGVGYDIVRGNPLGDPNLMGDPGLRSPVIRFSYSQDEEGVSSDLRELQP 94
R KT S G YLG GYD++RGNPLGD + DPG +S VI+ +S++ E +S+ LR LQP
Sbjct 341 RFKTPS-GLEYLGAGYDLIRGNPLGDSVTLLDPGYKSSVIQMHWSRNIENISNSLRFLQP 399
Query 95 RGAYSRPFVACKQSENLSEVSSLADYQKELEADASLVGGDSVGLNSFSASAAYRDIAKRT 154
G + RP+ +C + ++E++S K L ADAS+ + FSASA ++ + +
Sbjct 400 VGGWIRPYSSCHK---VTEINSCKSLLKSLSADASVSLSLPGDVFKFSASAKFKKLQDVS 456
Query 155 VKRNTKTFILKTYCLRFEAGLA 176
+K FI K+YC ++ AG++
Sbjct 457 KSGKSKMFINKSYCFKYVAGIS 478
> bbo:BBOV_II007150 18.m06592; mac/perforin domain containing
protein
Length=752
Score = 104 bits (259), Expect = 2e-22, Method: Composition-based stats.
Identities = 49/134 (36%), Positives = 78/134 (58%), Gaps = 0/134 (0%)
Query 42 GFCYLGVGYDIVRGNPLGDPNLMGDPGLRSPVIRFSYSQDEEGVSSDLRELQPRGAYSRP 101
G YLG GYD+++GNP+GD ++ DPG R+ V++ + D EG+S+ +QP+GA+ RP
Sbjct 421 GLEYLGAGYDLLKGNPMGDTIILLDPGYRASVVQMHWRDDAEGLSNSRHFIQPKGAWVRP 480
Query 102 FVACKQSENLSEVSSLADYQKELEADASLVGGDSVGLNSFSASAAYRDIAKRTVKRNTKT 161
+ +C + E +SEV+ L ADAS+ F+AS Y +I K + T
Sbjct 481 YTSCHKGETISEVAKTQSLDNVLSADASVSASLPGDKFKFAASVNYNNIKKAYDSKGVNT 540
Query 162 FILKTYCLRFEAGL 175
++ ++YC F AG+
Sbjct 541 YVSRSYCFNFVAGI 554
> tpv:TP03_0801 hypothetical protein
Length=353
Score = 90.1 bits (222), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 47/132 (35%), Positives = 73/132 (55%), Gaps = 2/132 (1%)
Query 45 YLGVGYDIVRGNPLGDPNLMGDPGLRSPVIRFSYSQDEEGVSSDLRELQPRGAYSRPFVA 104
YLG GYDI+ GNPL DP+L+ DPG R P+I +S +E + + A+ RP +
Sbjct 26 YLGCGYDILFGNPLSDPDLLVDPGFRDPIISYSIMFKKEKLFKKISYSNITNAWIRPLIE 85
Query 105 CKQSENLSEVSSLADYQKELEADASLVGGDSVGLNS-FSASAAYRDIAKRTVKRNTKTFI 163
CK+S + S V S+ Y+ + D+ +G S+ ++ FS S Y +I+ + K ++
Sbjct 86 CKRSNSRSVVDSMEKYKDIISVDSD-IGVSSIDESAKFSLSTNYSEISDLLKNNDNKLYV 144
Query 164 LKTYCLRFEAGL 175
K+YC EA L
Sbjct 145 DKSYCFLLEAAL 156
> tpv:TP04_0335 hypothetical protein
Length=441
Score = 89.7 bits (221), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 48/139 (34%), Positives = 79/139 (56%), Gaps = 3/139 (2%)
Query 23 AQQAVTMQDIYARAKTVSPGFCYLGVGYDIVRGNPLGDPNLMGDPGLRSPVIRFSYSQDE 82
+ V + I + + G YLG GYDIV+ N +GD + D G R+PVI F+++Q +
Sbjct 53 VEPIVENESINFKTTKIVVGLEYLGAGYDIVKANTMGDADQAEDLGYRAPVIDFTWAQTD 112
Query 83 EGVSSDLRELQPRGAYSRPFVACKQSENLSEVSSLADYQKELEADASL-VGGDSVGLNSF 141
GV++ L LQP G + RP V+C +SEN++E+ S++ + E+D + V +VG S
Sbjct 113 VGVTNSLDSLQPVGGWVRPKVSCGESENVTEIESISKLKDVTESDVGVNVKIPNVGTGSL 172
Query 142 SASAAYRDIAKRTVKRNTK 160
+ Y+++ K + N K
Sbjct 173 NTQ--YQELKKDSEHTNNK 189
> tpv:TP04_0337 hypothetical protein
Length=498
Score = 89.0 bits (219), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 43/131 (32%), Positives = 72/131 (54%), Gaps = 2/131 (1%)
Query 45 YLGVGYDIVRGNPLGDPNLMGDPGLRSPVIRFSYSQDEEGVSSDLRELQPRGAYSRPFVA 104
YLGVGYD + N +G + + DPG R+P+I F++ ++ EG S L L P G + RP +
Sbjct 17 YLGVGYDSIYANSVGSDSTLLDPGYRAPIIEFAWRKNSEGYSPTLGSLHPVGGWVRPVFS 76
Query 105 CKQSENLSEVSSLADYQKELEADASLVGGDSVGLNSFSASAAYRDIAKRTVKRNTKTFIL 164
C +S ++E+S+L + + L A L G + NSF+ S Y++ + K +
Sbjct 77 CSRSTKINEISNLEELKDSLSASTKLNG--DIPENSFTGSLEYKNALMNFKSKRQKIYNK 134
Query 165 KTYCLRFEAGL 175
C+R++ G+
Sbjct 135 TEQCVRYQVGI 145
> bbo:BBOV_III000410 hypothetical protein
Length=1272
Score = 87.8 bits (216), Expect = 2e-17, Method: Composition-based stats.
Identities = 46/115 (40%), Positives = 65/115 (56%), Gaps = 6/115 (5%)
Query 41 PGFCYLGVGYDIVRGNPLGDPNLMGDPGLRSPVIRFSYSQDEEGVSSDLRELQPRGAYSR 100
P YLG GYDI+ GNPL D + DPG R+P+I F+ + + DL+ GA+ R
Sbjct 942 PAIEYLGCGYDILYGNPLADDGTLVDPGYRNPIISFTLAHHKSKGKKDLKYANIPGAWIR 1001
Query 101 PFVACKQSENLSEVSSLADYQKELEADASLVGGDSVGLNSFSASAAYRDIAKRTV 155
P VAC++S+ S V S++DYQK L D+ VG+ + SA + A+ TV
Sbjct 1002 PLVACQRSDETSIVKSISDYQKALSVDS------EVGIGTVDESAKFALSAEITV 1050
Score = 35.4 bits (80), Expect = 0.11, Method: Composition-based stats.
Identities = 15/37 (40%), Positives = 18/37 (48%), Gaps = 0/37 (0%)
Query 42 GFCYLGVGYDIVRGNPLGDPNLMGDPGLRSPVIRFSY 78
G YLG GYD + P GD D G PV+ F +
Sbjct 136 GLEYLGCGYDSTKSMPFGDEESFLDSGYTQPVVNFQW 172
> bbo:BBOV_II002020 18.m06160; mac/perforin domain containing
protein
Length=420
Score = 85.9 bits (211), Expect = 7e-17, Method: Compositional matrix adjust.
Identities = 40/118 (33%), Positives = 70/118 (59%), Gaps = 2/118 (1%)
Query 40 SPGFCYLGVGYDIVRGNPLGDPNLMGDPGLRSPVIRFSYSQDEEGVSSDLRELQPRGAYS 99
+PG YLG+GYD ++GN +G + DPG R+P+I F++ + EG S L + P +
Sbjct 75 TPGMDYLGIGYDAIKGNTMGGEESLLDPGYRAPIINFNWRKSAEGYSPSLNAVYPLYGWV 134
Query 100 RPFVACKQSENLSEVSSLADYQKELEADASLVGGDSVGLNSFSASAAYRDIAKRTVKR 157
RP +C +S + E+ +L + +K A+AS+ G + +FSASA Y++ +++ +
Sbjct 135 RPVYSCGRSSKIQEIENLDELKKVFSANASIKG--DIPAVAFSASAKYKNASEKLAHK 190
> bbo:BBOV_II001970 18.m09950; mac/perforin domain containing
membrane protein
Length=559
Score = 82.8 bits (203), Expect = 5e-16, Method: Composition-based stats.
Identities = 50/146 (34%), Positives = 80/146 (54%), Gaps = 6/146 (4%)
Query 32 IYARAKTVSPGFCYLGVGYDIVRGNPLGDPNLMGDPGLRSPVIRFSYSQDEEGVSSDLRE 91
I++ + VS G YLG GYD V+ + L N D G RSP++ F +++ + GV++ L+
Sbjct 49 IFSADRIVS-GLEYLGSGYDAVKASGLVSINNGDDLGHRSPIVDFYWAKSDVGVTNSLKW 107
Query 92 LQPRGAYSRPFVACKQSENLSEVSSLADYQKELEADASLVGG--DSVGLNSFSASAAYRD 149
LQP G + RP AC +SE ++ S + ++ + D GG S GL S + A Y D
Sbjct 108 LQPLGGWVRPITACGESETVTVGSQQSTNEESTQFD---FGGFAMSSGLGSGALRAGYTD 164
Query 150 IAKRTVKRNTKTFILKTYCLRFEAGL 175
I+ +T ++K + YC + AG+
Sbjct 165 ISGKTQHISSKQYTNSYYCFTYAAGM 190
> pfa:PFL0805w MAC/Perforin, putative
Length=1073
Score = 53.9 bits (128), Expect = 3e-07, Method: Composition-based stats.
Identities = 29/89 (32%), Positives = 50/89 (56%), Gaps = 5/89 (5%)
Query 91 ELQPRGAYSRPFV----ACKQSENLSEVSSLADYQKELEADASLVGGDSVGLNSFSASAA 146
++Q +P+V +C QS+N+ E+ +L Y+ EL +D + S SFSASA
Sbjct 511 KIQTTNESMKPWVIPEHSCSQSKNVEEIRNLEQYKLELLSDVKVSTPSSFPY-SFSASAE 569
Query 147 YRDIAKRTVKRNTKTFILKTYCLRFEAGL 175
+++ K+ +N F++K YCLR+ G+
Sbjct 570 FKNALKKLKVQNNVIFLMKIYCLRYYTGI 598
Score = 53.9 bits (128), Expect = 3e-07, Method: Composition-based stats.
Identities = 25/68 (36%), Positives = 38/68 (55%), Gaps = 9/68 (13%)
Query 13 EERRKQEWEAAQQAVTMQDIYARAKTVSPGFCYLGVGYDIVRGNPLGDPNLMGDPGLRSP 72
+E+RK+ ++++Y K YLG+GYDI+ GNP GDP L DPG R P
Sbjct 381 KEKRKE---------NLKEVYNDKKNNYLSLKYLGLGYDIIMGNPEGDPTLNVDPGFRGP 431
Query 73 VIRFSYSQ 80
V++ + +
Sbjct 432 VLQINLKE 439
> pfa:PF08_0052 perforin like protein 5
Length=676
Score = 52.0 bits (123), Expect = 1e-06, Method: Composition-based stats.
Identities = 36/127 (28%), Positives = 62/127 (48%), Gaps = 5/127 (3%)
Query 45 YLGVGYDIVRGNPLGDPNLMGDPGLRSPVIRFSYSQDEEGVSSDLRELQPRGAYSRPFVA 104
YLG+ YDI++GNP GDP + D G R V++ + + D+ + + G S+ +
Sbjct 49 YLGMSYDIIKGNPWGDPIYVIDLGYRRNVLKKRKLNSDNNIKDDIVKFKV-GEASK--IK 105
Query 105 CKQSENLSEVSSLADYQKELEADASLVGGDSVGLNSFSASAAYRDIAKRTVKRNTKTFIL 164
C + + + +L D KE E S V + ++ F+ S Y+ + KR + R I
Sbjct 106 CIDTIKENVIDNLCDINKEYERSYS-VSSINDDIHPFNDSNYYKMLVKR-INRGDSIIIE 163
Query 165 KTYCLRF 171
K C ++
Sbjct 164 KKLCSKY 170
> tpv:TP03_0810 hypothetical protein
Length=348
Score = 46.6 bits (109), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 40/132 (30%), Positives = 60/132 (45%), Gaps = 8/132 (6%)
Query 45 YLGVGYDIVRGNPLGDPNLMGDPGLRSPVIRFSYSQDEEGVSSDLRELQPRGAYSRPFVA 104
++G G+D+V GNPL N + G RSP+I Y + G ++ + G + R
Sbjct 38 HVGFGFDLVEGNPLDSFNDLNTFGFRSPIIVQPYITRDIG---NIIIKRNNGIWVRKSNN 94
Query 105 CKQSENLSEVSSLADYQKELEADASLVGGDSVGL-NSFSASAAYRDIAKRTVKRNTKTFI 163
C Q+ ++ +D +EL D SL S L N S DI +K I
Sbjct 95 CTQNYEPRDIERGSDLVRELFNDFSLDSPFSEELWNRNSKGLGLNDITFNK----SKFKI 150
Query 164 LKTYCLRFEAGL 175
+K YC +E+GL
Sbjct 151 VKCYCSLYESGL 162
> bbo:BBOV_III000320 17.m10445; hypothetical protein
Length=512
Score = 38.9 bits (89), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 34/129 (26%), Positives = 59/129 (45%), Gaps = 9/129 (6%)
Query 48 VGYDIVRGNPLGDPNLMGDPGLRSPVIRFSYSQDEEGVSSD-LRELQPRGAYSRPFVACK 106
+GYD V GNP G D G R+P++ + +S D + + G + R C
Sbjct 1 MGYDAVLGNPFGSLGQDKDSGYRNPIL-----ETHVTISGDKTKSSEQNGLWVRELSTCW 55
Query 107 QSENLSEVSSLADYQKELEADASLVGGDSVGLNSFSASAAYRDIAKRTVKRNTKTFILKT 166
S+ +V + +EL+ + ++ G ++ L S S ++ D K T K I K+
Sbjct 56 ISDTHDDVGD-DELVRELQNEFTVEGSENSELLSASINSMADD--KPTSKHTVNYRIAKS 112
Query 167 YCLRFEAGL 175
+C E+G+
Sbjct 113 FCAIRESGI 121
> pfa:PF08_0050 MAC/Perforin, putative
Length=654
Score = 37.0 bits (84), Expect = 0.036, Method: Composition-based stats.
Identities = 21/52 (40%), Positives = 26/52 (50%), Gaps = 0/52 (0%)
Query 30 QDIYARAKTVSPGFCYLGVGYDIVRGNPLGDPNLMGDPGLRSPVIRFSYSQD 81
DI KT YLG GYDI+ G PL + L+ DPG + +I S D
Sbjct 28 HDIKTDEKTNEIFSKYLGKGYDILFGYPLPNNELIDDPGFKEVIIDTQLSID 79
> cel:C54D2.5 cca-1; Calcium Channel, Alpha subunit family member
(cca-1); K05315 voltage-dependent calcium channel alpha
1, invertebrate
Length=1844
Score = 31.6 bits (70), Expect = 1.4, Method: Composition-based stats.
Identities = 17/41 (41%), Positives = 24/41 (58%), Gaps = 6/41 (14%)
Query 9 EARREERRKQEWEAAQQAVTMQDIYAR------AKTVSPGF 43
E++ EE+R+ E +A +QAV +D R AKT SP F
Sbjct 891 ESKEEEKRQLEEDARKQAVEEEDERKRELELIIAKTTSPAF 931
> tpv:TP03_0798 hypothetical protein
Length=357
Score = 30.4 bits (67), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 21/68 (30%), Positives = 30/68 (44%), Gaps = 9/68 (13%)
Query 42 GFCYLGVGYDIVRGNPLG----DPNLMGDPGLRSPVIRFSYSQDEEGVSSDLRELQPRGA 97
G YLG+GY+I +P G DP D + PVI F + G +++ P G
Sbjct 39 GLEYLGLGYNIKESSPFGNDEDDP---IDLAYKEPVISFENATSLVGKGAEIN--LPLGV 93
Query 98 YSRPFVAC 105
+ R C
Sbjct 94 WIRNESLC 101
> sce:YBR170C NPL4, HRD4; Endoplasmic reticulum and nuclear membrane
protein, forms a complex with Cdc48p and Ufd1p that recognizes
ubiquitinated proteins in the endoplasmic reticulum
and delivers them to the proteasome for degradation; K14015
nuclear protein localization protein 4 homolog
Length=580
Score = 30.4 bits (67), Expect = 3.5, Method: Composition-based stats.
Identities = 19/55 (34%), Positives = 28/55 (50%), Gaps = 1/55 (1%)
Query 95 RGAYSRPFVACKQSENLSEVSSLADYQKELEADASLVGGDSVGLNSFSASAAYRD 149
+G +S FV C S NL ++ YQ EA+A LV D + ++F + A D
Sbjct 374 QGFFSSKFVTCVISGNLEGEIDISSYQVSTEAEA-LVTADMISGSTFPSMAYIND 427
> dre:100003644 fc38h03; wu:fc38h03
Length=415
Score = 30.0 bits (66), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 16/40 (40%), Positives = 21/40 (52%), Gaps = 2/40 (5%)
Query 59 GDPNLMGDPGLRSPVIRFSYS--QDEEGVSSDLRELQPRG 96
GDP LMG PG+R PV + + E+G D P+G
Sbjct 160 GDPGLMGMPGMRGPVGPKGLAGYKGEKGARGDFGPAGPKG 199
Lambda K H
0.315 0.131 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4600750868
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40