bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2196_orf1
Length=102
Score E
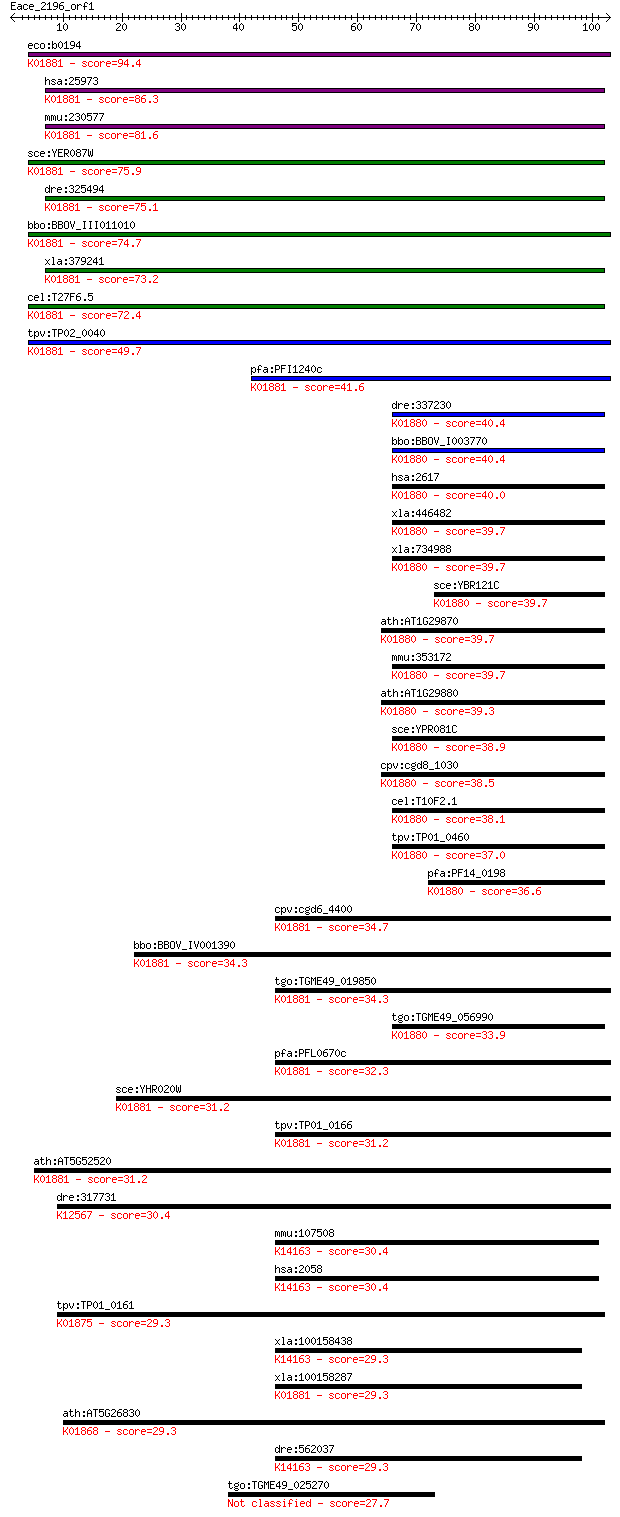
Sequences producing significant alignments: (Bits) Value
eco:b0194 proS, drpA, ECK0194, JW0190; prolyl-tRNA synthetase ... 94.4 9e-20
hsa:25973 PARS2, DKFZp727A071, MGC14416, MGC19467, MT-PRORS; p... 86.3 2e-17
mmu:230577 Pars2, BC027073, MGC47008; prolyl-tRNA synthetase (... 81.6 5e-16
sce:YER087W AIM10; Aim10p (EC:6.1.1.15); K01881 prolyl-tRNA sy... 75.9 3e-14
dre:325494 pars2, fc83e06, wu:fc83e06, zgc:77163; prolyl-tRNA ... 75.1 5e-14
bbo:BBOV_III011010 17.m07948; tRNA synthetase class II core do... 74.7 6e-14
xla:379241 pars2, MGC53152; prolyl-tRNA synthetase 2, mitochon... 73.2 2e-13
cel:T27F6.5 prs-2; Prolyl tRNA Synthetase family member (prs-2... 72.4 3e-13
tpv:TP02_0040 prolyl-tRNA synthetase; K01881 prolyl-tRNA synth... 49.7 2e-06
pfa:PFI1240c prolyl-t-RNA synthase, putative (EC:6.1.1.15); K0... 41.6 7e-04
dre:337230 fb02f03, wu:fb02f03; si:dkey-276i5.1; K01880 glycyl... 40.4 0.001
bbo:BBOV_I003770 19.m02173; glycyl-tRNA synthetase (EC:6.1.1.1... 40.4 0.002
hsa:2617 GARS, CMT2D, DSMAV, GlyRS, HMN5, SMAD1; glycyl-tRNA s... 40.0 0.002
xla:446482 gars, MGC79076; glycyl-tRNA synthetase (EC:6.1.1.14... 39.7 0.002
xla:734988 hypothetical protein MGC130901; K01880 glycyl-tRNA ... 39.7 0.002
sce:YBR121C GRS1; Grs1p (EC:6.1.1.14); K01880 glycyl-tRNA synt... 39.7 0.002
ath:AT1G29870 tRNA synthetase class II (G, H, P and S) family ... 39.7 0.002
mmu:353172 Gars, Nmf249, Sgrp23; glycyl-tRNA synthetase (EC:6.... 39.7 0.002
ath:AT1G29880 glycyl-tRNA synthetase / glycine--tRNA ligase (E... 39.3 0.003
sce:YPR081C GRS2; Grs2p (EC:6.1.1.14); K01880 glycyl-tRNA synt... 38.9 0.004
cpv:cgd8_1030 glycyl-tRNA synthetase ; K01880 glycyl-tRNA synt... 38.5 0.005
cel:T10F2.1 grs-1; Glycyl tRNA Synthetase family member (grs-1... 38.1 0.006
tpv:TP01_0460 glycyl-tRNA synthetase; K01880 glycyl-tRNA synth... 37.0 0.017
pfa:PF14_0198 glycine-tRNA ligase, putative; K01880 glycyl-tRN... 36.6 0.019
cpv:cgd6_4400 proline-tRNA synthetase; class II aaRS (ybak RNA... 34.7 0.082
bbo:BBOV_IV001390 21.m02889; prolyl-tRNA synthetase (EC:6.1.1.... 34.3 0.087
tgo:TGME49_019850 prolyl-tRNA synthetase, putative (EC:6.1.1.1... 34.3 0.092
tgo:TGME49_056990 glycyl-tRNA synthetase, putative (EC:6.1.1.1... 33.9 0.13
pfa:PFL0670c bifunctional aminoacyl-tRNA synthetase, putative ... 32.3 0.35
sce:YHR020W Protein of unknown function that may interact with... 31.2 0.73
tpv:TP01_0166 prolyl-tRNA synthetase; K01881 prolyl-tRNA synth... 31.2 0.75
ath:AT5G52520 OVA6; OVA6 (OVULE ABORTION 6); ATP binding / ami... 31.2 0.90
dre:317731 ttna, fi20g08, si:busm1-258d18.1, si:dz258d18.1, tt... 30.4 1.3
mmu:107508 Eprs, 2410081F06Rik, 3010002K18Rik, C79379, Qprs; g... 30.4 1.3
hsa:2058 EPRS, DKFZp313B047, EARS, GLUPRORS, PARS, QARS, QPRS;... 30.4 1.5
tpv:TP01_0161 seryl-tRNA synthetase; K01875 seryl-tRNA synthet... 29.3 2.9
xla:100158438 eprs; glutamyl-prolyl-tRNA synthetase; K14163 bi... 29.3 3.2
xla:100158287 hypothetical protein LOC100158287; K01881 prolyl... 29.3 3.3
ath:AT5G26830 threonyl-tRNA synthetase / threonine--tRNA ligas... 29.3 3.3
dre:562037 eprs, wu:fb38d08, wu:ft34d10; glutamyl-prolyl-tRNA ... 29.3 3.5
tgo:TGME49_025270 hypothetical protein 27.7 8.2
> eco:b0194 proS, drpA, ECK0194, JW0190; prolyl-tRNA synthetase
(EC:6.1.1.15); K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=572
Score = 94.4 bits (233), Expect = 9e-20, Method: Composition-based stats.
Identities = 46/99 (46%), Positives = 59/99 (59%), Gaps = 0/99 (0%)
Query 4 ASPLSLPNIMPIHWLQNSDRIRSFGPSLYRLQDRRGRQFFLPPTSEEAAAWLVASNIRSH 63
A +S+P + P Q S R +GP L R DR R F L PT EE L+ + + S+
Sbjct 66 AIEVSMPVVQPADLWQESGRWEQYGPELLRFVDRGERPFVLGPTHEEVITDLIRNELSSY 125
Query 64 RDLPQIFYQIGPKFRDEARPRAGCLRAREFVMLEVYSFH 102
+ LP FYQI KFRDE RPR G +R+REF+M + YSFH
Sbjct 126 KQLPLNFYQIQTKFRDEVRPRFGVMRSREFLMKDAYSFH 164
> hsa:25973 PARS2, DKFZp727A071, MGC14416, MGC19467, MT-PRORS;
prolyl-tRNA synthetase 2, mitochondrial (putative) (EC:6.1.1.15);
K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=475
Score = 86.3 bits (212), Expect = 2e-17, Method: Composition-based stats.
Identities = 42/96 (43%), Positives = 62/96 (64%), Gaps = 1/96 (1%)
Query 7 LSLPNIMPIHWLQNSDRIRSFGPSLYRLQDRRGRQFFLPPTSEEAAAWLVASNIR-SHRD 65
+++P++ P Q ++R G L RL+DR G+++ L PT EEA L+AS + S++
Sbjct 117 VNMPSLSPAELWQATNRWDLMGKELLRLRDRHGKEYCLGPTHEEAITALIASQKKLSYKQ 176
Query 66 LPQIFYQIGPKFRDEARPRAGCLRAREFVMLEVYSF 101
LP + YQ+ KFRDE RPR G LR REF M ++Y+F
Sbjct 177 LPFLLYQVTRKFRDEPRPRFGLLRGREFYMKDMYTF 212
> mmu:230577 Pars2, BC027073, MGC47008; prolyl-tRNA synthetase
(mitochondrial)(putative) (EC:6.1.1.15); K01881 prolyl-tRNA
synthetase [EC:6.1.1.15]
Length=511
Score = 81.6 bits (200), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 42/96 (43%), Positives = 61/96 (63%), Gaps = 1/96 (1%)
Query 7 LSLPNIMPIHWLQNSDRIRSFGPSLYRLQDRRGRQFFLPPTSEEAAAWLVASNIR-SHRD 65
+++P++ P + + R G L RL+DR G+++ L PT EEA LVAS + S++
Sbjct 153 INMPSLSPAELWRATSRWDLMGRELLRLRDRHGKEYCLGPTHEEAVTALVASQKKLSYKQ 212
Query 66 LPQIFYQIGPKFRDEARPRAGCLRAREFVMLEVYSF 101
LP + YQ+ KFRDE RPR G LR REF M ++Y+F
Sbjct 213 LPLLLYQVTRKFRDEPRPRFGLLRGREFYMKDMYTF 248
> sce:YER087W AIM10; Aim10p (EC:6.1.1.15); K01881 prolyl-tRNA
synthetase [EC:6.1.1.15]
Length=576
Score = 75.9 bits (185), Expect = 3e-14, Method: Composition-based stats.
Identities = 40/98 (40%), Positives = 58/98 (59%), Gaps = 2/98 (2%)
Query 4 ASPLSLPNIMPIHWLQNSDRIRSFGPSLYRLQDRRGRQFFLPPTSEEAAAWLVASNIRSH 63
A +SL I Q +DR + L++L+D +G+Q+ L T EE L+ + I S+
Sbjct 74 AIEVSLSAISSKALWQATDRWNN--SELFKLKDSKGKQYCLTATCEEDITDLMKNYIASY 131
Query 64 RDLPQIFYQIGPKFRDEARPRAGCLRAREFVMLEVYSF 101
+D+P YQ+ K+RDE RPR G LR REF+M + YSF
Sbjct 132 KDMPITIYQMTRKYRDEIRPRGGILRGREFLMKDAYSF 169
> dre:325494 pars2, fc83e06, wu:fc83e06, zgc:77163; prolyl-tRNA
synthetase (mitochondrial) (EC:6.1.1.15); K01881 prolyl-tRNA
synthetase [EC:6.1.1.15]
Length=477
Score = 75.1 bits (183), Expect = 5e-14, Method: Composition-based stats.
Identities = 38/96 (39%), Positives = 54/96 (56%), Gaps = 1/96 (1%)
Query 7 LSLPNIMPIHWLQNSDRIRSFGPSLYRLQDRRGRQFFLPPTSEEAAAWLVASNIR-SHRD 65
+ +P + + S R G ++RL DR ++ L PT EEA L AS S++
Sbjct 118 VDMPALCSAELWKKSGRWELMGKEMFRLLDRHNGEYCLGPTHEEAVTELFASQTTMSYKK 177
Query 66 LPQIFYQIGPKFRDEARPRAGCLRAREFVMLEVYSF 101
LP + YQ+ KFRDE +P+ G LR REF M ++YSF
Sbjct 178 LPLLLYQVTRKFRDEQKPKFGLLRGREFYMKDMYSF 213
> bbo:BBOV_III011010 17.m07948; tRNA synthetase class II core
domain containing protein; K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=439
Score = 74.7 bits (182), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 41/101 (40%), Positives = 57/101 (56%), Gaps = 4/101 (3%)
Query 4 ASPLSLPNIMPIHWLQNSDRIRSFGPSLYRLQDRRGRQFFLPPTSEEAAAWLVASNIR-- 61
A + LP ++ + ++R FG L+ L DR G+ + L PT EE ++ +
Sbjct 88 AKEVELPMLLSSDTI--NERRNEFGRELFSLTDRNGKHYDLCPTCEELICRMLDKALSPL 145
Query 62 SHRDLPQIFYQIGPKFRDEARPRAGCLRAREFVMLEVYSFH 102
S R+LP YQIG KFRDEARP +R REF+M + YSFH
Sbjct 146 SKRNLPIYLYQIGRKFRDEARPCNSNMRCREFIMKDAYSFH 186
> xla:379241 pars2, MGC53152; prolyl-tRNA synthetase 2, mitochondrial
(putative); K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=466
Score = 73.2 bits (178), Expect = 2e-13, Method: Composition-based stats.
Identities = 39/96 (40%), Positives = 56/96 (58%), Gaps = 1/96 (1%)
Query 7 LSLPNIMPIHWLQNSDRIRSFGPSLYRLQDRRGRQFFLPPTSEEAAAWLVASNIR-SHRD 65
+ +P + + S R G L+RL+DR +++ L PT EE LVAS ++++
Sbjct 108 IDMPTLGSAALWRQSGRWDLMGKELFRLKDRHNQEYCLGPTHEELVTHLVASEGGINNKE 167
Query 66 LPQIFYQIGPKFRDEARPRAGCLRAREFVMLEVYSF 101
LP + YQI KFRDE RP G LR REF M ++Y+F
Sbjct 168 LPLLLYQITRKFRDEQRPCFGLLRGREFYMKDMYTF 203
> cel:T27F6.5 prs-2; Prolyl tRNA Synthetase family member (prs-2);
K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=454
Score = 72.4 bits (176), Expect = 3e-13, Method: Composition-based stats.
Identities = 36/99 (36%), Positives = 54/99 (54%), Gaps = 1/99 (1%)
Query 4 ASPLSLPNIMPIHWLQNSDRIRSFGPSLYRLQDRRGRQFFLPPTSEEAAAWLVAS-NIRS 62
A +++P + + + R + GP + R DR+ L PT+EE L+A+ +
Sbjct 66 ALKIAMPIVGTQQLWEKTGRWEAMGPEMIRFHDRQQNHMCLQPTAEEMCTELIATLSPLK 125
Query 63 HRDLPQIFYQIGPKFRDEARPRAGCLRAREFVMLEVYSF 101
P I YQIG KFRDE PR G +R R+F+M ++YSF
Sbjct 126 KSQFPLIVYQIGEKFRDEMNPRFGLMRGRQFLMKDMYSF 164
> tpv:TP02_0040 prolyl-tRNA synthetase; K01881 prolyl-tRNA synthetase
[EC:6.1.1.15]
Length=461
Score = 49.7 bits (117), Expect = 2e-06, Method: Composition-based stats.
Identities = 31/101 (30%), Positives = 51/101 (50%), Gaps = 3/101 (2%)
Query 4 ASPLSLPNIMPIHWLQNSDRIRSFGPSLYRLQDRRGRQFFLPPTSEEAAAWLVASNI--R 61
A+ LS P + P + N+ L+++ ++ L T E+ ++ + +
Sbjct 106 AAQLSFPLLNPEKVVSNTGNT-VLDNDLFKVSGNGSVEYRLSGTCEDLCNIVLKNEMIPL 164
Query 62 SHRDLPQIFYQIGPKFRDEARPRAGCLRAREFVMLEVYSFH 102
S LP + YQI KFRDE R G +R +EF+ML++YS H
Sbjct 165 SKLQLPVLLYQINTKFRDELRVNEGKVRLKEFLMLDMYSLH 205
> pfa:PFI1240c prolyl-t-RNA synthase, putative (EC:6.1.1.15);
K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=579
Score = 41.6 bits (96), Expect = 7e-04, Method: Composition-based stats.
Identities = 23/65 (35%), Positives = 37/65 (56%), Gaps = 5/65 (7%)
Query 42 FFLPPTSEEAAAWLVAS----NIRSHRDLPQIFYQIGPKFRDEARPRAGCLRAREFVMLE 97
+ L PT EE++ L+ NI + + LP + +Q KFR+E R +++EF+M +
Sbjct 178 YILSPTCEESSLSLINQIYNENI-TIKCLPLLIHQYNYKFRNEKRFEKSLFKSKEFLMKD 236
Query 98 VYSFH 102
YSFH
Sbjct 237 GYSFH 241
> dre:337230 fb02f03, wu:fb02f03; si:dkey-276i5.1; K01880 glycyl-tRNA
synthetase [EC:6.1.1.14]
Length=764
Score = 40.4 bits (93), Expect = 0.001, Method: Composition-based stats.
Identities = 18/36 (50%), Positives = 22/36 (61%), Gaps = 0/36 (0%)
Query 66 LPQIFYQIGPKFRDEARPRAGCLRAREFVMLEVYSF 101
LP QIG FR+E PR+G +R REF M E+ F
Sbjct 344 LPFAAAQIGNSFRNEISPRSGLIRVREFTMAEIEHF 379
> bbo:BBOV_I003770 19.m02173; glycyl-tRNA synthetase (EC:6.1.1.14);
K01880 glycyl-tRNA synthetase [EC:6.1.1.14]
Length=733
Score = 40.4 bits (93), Expect = 0.002, Method: Composition-based stats.
Identities = 19/36 (52%), Positives = 22/36 (61%), Gaps = 0/36 (0%)
Query 66 LPQIFYQIGPKFRDEARPRAGCLRAREFVMLEVYSF 101
+P QIG FR+E PR G LR REF+M EV F
Sbjct 318 MPLAAAQIGLGFRNEISPRNGLLRVREFLMAEVEYF 353
> hsa:2617 GARS, CMT2D, DSMAV, GlyRS, HMN5, SMAD1; glycyl-tRNA
synthetase (EC:6.1.1.14); K01880 glycyl-tRNA synthetase [EC:6.1.1.14]
Length=739
Score = 40.0 bits (92), Expect = 0.002, Method: Composition-based stats.
Identities = 18/36 (50%), Positives = 22/36 (61%), Gaps = 0/36 (0%)
Query 66 LPQIFYQIGPKFRDEARPRAGCLRAREFVMLEVYSF 101
LP QIG FR+E PR+G +R REF M E+ F
Sbjct 319 LPFAAAQIGNSFRNEISPRSGLIRVREFTMAEIEHF 354
> xla:446482 gars, MGC79076; glycyl-tRNA synthetase (EC:6.1.1.14);
K01880 glycyl-tRNA synthetase [EC:6.1.1.14]
Length=747
Score = 39.7 bits (91), Expect = 0.002, Method: Composition-based stats.
Identities = 18/36 (50%), Positives = 22/36 (61%), Gaps = 0/36 (0%)
Query 66 LPQIFYQIGPKFRDEARPRAGCLRAREFVMLEVYSF 101
LP QIG FR+E PR+G +R REF M E+ F
Sbjct 327 LPFAAAQIGNSFRNEISPRSGLIRVREFTMAEIEHF 362
> xla:734988 hypothetical protein MGC130901; K01880 glycyl-tRNA
synthetase [EC:6.1.1.14]
Length=740
Score = 39.7 bits (91), Expect = 0.002, Method: Composition-based stats.
Identities = 18/36 (50%), Positives = 22/36 (61%), Gaps = 0/36 (0%)
Query 66 LPQIFYQIGPKFRDEARPRAGCLRAREFVMLEVYSF 101
LP QIG FR+E PR+G +R REF M E+ F
Sbjct 320 LPFAAAQIGNSFRNEISPRSGLIRVREFTMAEIEHF 355
> sce:YBR121C GRS1; Grs1p (EC:6.1.1.14); K01880 glycyl-tRNA synthetase
[EC:6.1.1.14]
Length=667
Score = 39.7 bits (91), Expect = 0.002, Method: Composition-based stats.
Identities = 17/29 (58%), Positives = 20/29 (68%), Gaps = 0/29 (0%)
Query 73 IGPKFRDEARPRAGCLRAREFVMLEVYSF 101
IG FR+E PRAG LR REF+M E+ F
Sbjct 255 IGKSFRNEISPRAGLLRVREFLMAEIEHF 283
> ath:AT1G29870 tRNA synthetase class II (G, H, P and S) family
protein; K01880 glycyl-tRNA synthetase [EC:6.1.1.14]
Length=463
Score = 39.7 bits (91), Expect = 0.002, Method: Composition-based stats.
Identities = 18/38 (47%), Positives = 22/38 (57%), Gaps = 0/38 (0%)
Query 64 RDLPQIFYQIGPKFRDEARPRAGCLRAREFVMLEVYSF 101
R LP Q+G FR+E PR G LR REF + E+ F
Sbjct 257 RKLPFAVAQVGRVFRNEISPRQGLLRTREFTLAEIEHF 294
> mmu:353172 Gars, Nmf249, Sgrp23; glycyl-tRNA synthetase (EC:6.1.1.14);
K01880 glycyl-tRNA synthetase [EC:6.1.1.14]
Length=729
Score = 39.7 bits (91), Expect = 0.002, Method: Composition-based stats.
Identities = 18/36 (50%), Positives = 22/36 (61%), Gaps = 0/36 (0%)
Query 66 LPQIFYQIGPKFRDEARPRAGCLRAREFVMLEVYSF 101
LP QIG FR+E PR+G +R REF M E+ F
Sbjct 309 LPFAAAQIGNSFRNEISPRSGLIRVREFTMAEIEHF 344
> ath:AT1G29880 glycyl-tRNA synthetase / glycine--tRNA ligase
(EC:6.1.1.14); K01880 glycyl-tRNA synthetase [EC:6.1.1.14]
Length=729
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 18/38 (47%), Positives = 22/38 (57%), Gaps = 0/38 (0%)
Query 64 RDLPQIFYQIGPKFRDEARPRAGCLRAREFVMLEVYSF 101
+ LP QIG FR+E PR G LR REF + E+ F
Sbjct 314 KKLPFAAAQIGQAFRNEISPRQGLLRVREFTLAEIEHF 351
> sce:YPR081C GRS2; Grs2p (EC:6.1.1.14); K01880 glycyl-tRNA synthetase
[EC:6.1.1.14]
Length=618
Score = 38.9 bits (89), Expect = 0.004, Method: Composition-based stats.
Identities = 17/36 (47%), Positives = 22/36 (61%), Gaps = 0/36 (0%)
Query 66 LPQIFYQIGPKFRDEARPRAGCLRAREFVMLEVYSF 101
+P IG FR+E PR+G LR REF+M E+ F
Sbjct 207 IPFASASIGKSFRNEISPRSGLLRVREFLMAEIEHF 242
> cpv:cgd8_1030 glycyl-tRNA synthetase ; K01880 glycyl-tRNA synthetase
[EC:6.1.1.14]
Length=656
Score = 38.5 bits (88), Expect = 0.005, Method: Composition-based stats.
Identities = 19/38 (50%), Positives = 22/38 (57%), Gaps = 0/38 (0%)
Query 64 RDLPQIFYQIGPKFRDEARPRAGCLRAREFVMLEVYSF 101
+ LP QIG FR+E PR G LR REF M E+ F
Sbjct 214 KKLPFSVAQIGLGFRNEIAPRNGLLRVREFQMAEIEHF 251
> cel:T10F2.1 grs-1; Glycyl tRNA Synthetase family member (grs-1);
K01880 glycyl-tRNA synthetase [EC:6.1.1.14]
Length=742
Score = 38.1 bits (87), Expect = 0.006, Method: Composition-based stats.
Identities = 18/36 (50%), Positives = 21/36 (58%), Gaps = 0/36 (0%)
Query 66 LPQIFYQIGPKFRDEARPRAGCLRAREFVMLEVYSF 101
LP QIG FR+E PR G +R REF M E+ F
Sbjct 329 LPFAAAQIGLGFRNEISPRQGLIRVREFTMCEIEHF 364
> tpv:TP01_0460 glycyl-tRNA synthetase; K01880 glycyl-tRNA synthetase
[EC:6.1.1.14]
Length=874
Score = 37.0 bits (84), Expect = 0.017, Method: Composition-based stats.
Identities = 18/36 (50%), Positives = 21/36 (58%), Gaps = 0/36 (0%)
Query 66 LPQIFYQIGPKFRDEARPRAGCLRAREFVMLEVYSF 101
+P QIG FR+E PR G LR REF M E+ F
Sbjct 429 IPFAAAQIGLGFRNEISPRNGLLRVREFPMAEIEYF 464
> pfa:PF14_0198 glycine-tRNA ligase, putative; K01880 glycyl-tRNA
synthetase [EC:6.1.1.14]
Length=889
Score = 36.6 bits (83), Expect = 0.019, Method: Composition-based stats.
Identities = 16/30 (53%), Positives = 19/30 (63%), Gaps = 0/30 (0%)
Query 72 QIGPKFRDEARPRAGCLRAREFVMLEVYSF 101
Q+G FR+E PR G LR REF M E+ F
Sbjct 472 QLGLGFRNEISPRNGLLRVREFQMAEIEYF 501
> cpv:cgd6_4400 proline-tRNA synthetase; class II aaRS (ybak RNA
binding domain plus tRNA synthetase) ; K01881 prolyl-tRNA
synthetase [EC:6.1.1.15]
Length=719
Score = 34.7 bits (78), Expect = 0.082, Method: Composition-based stats.
Identities = 22/57 (38%), Positives = 27/57 (47%), Gaps = 0/57 (0%)
Query 46 PTSEEAAAWLVASNIRSHRDLPQIFYQIGPKFRDEARPRAGCLRAREFVMLEVYSFH 102
PTSE A IRSHRDLP Q R E + +R REF+ E ++ H
Sbjct 330 PTSETIMYPYFAKWIRSHRDLPLKINQWTSIVRWEFKHPTPFIRTREFLWQEGHTAH 386
> bbo:BBOV_IV001390 21.m02889; prolyl-tRNA synthetase (EC:6.1.1.15);
K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=705
Score = 34.3 bits (77), Expect = 0.087, Method: Composition-based stats.
Identities = 27/85 (31%), Positives = 36/85 (42%), Gaps = 4/85 (4%)
Query 22 DRIRSFGPSLYRLQDRRGRQFFLP----PTSEEAAAWLVASNIRSHRDLPQIFYQIGPKF 77
+ I F P + + F P PTSE + IRSHRDLP Q
Sbjct 292 NHIEGFSPEVAWVTKYGDSDFVEPIAIRPTSETIMYPEFSKWIRSHRDLPLKLNQWCSVV 351
Query 78 RDEARPRAGCLRAREFVMLEVYSFH 102
R E + LR+REF+ E ++ H
Sbjct 352 RWEFKQPTPFLRSREFLWQEGHTAH 376
> tgo:TGME49_019850 prolyl-tRNA synthetase, putative (EC:6.1.1.15);
K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=711
Score = 34.3 bits (77), Expect = 0.092, Method: Composition-based stats.
Identities = 23/57 (40%), Positives = 27/57 (47%), Gaps = 0/57 (0%)
Query 46 PTSEEAAAWLVASNIRSHRDLPQIFYQIGPKFRDEARPRAGCLRAREFVMLEVYSFH 102
PTSE A IRSHRDLP Q R E + LR REF+ E ++ H
Sbjct 319 PTSETIMYPAYAKWIRSHRDLPLKLNQWCSVVRWEFKQPTPFLRTREFLWQEGHTAH 375
> tgo:TGME49_056990 glycyl-tRNA synthetase, putative (EC:6.1.1.14
6.1.1.3); K01880 glycyl-tRNA synthetase [EC:6.1.1.14]
Length=771
Score = 33.9 bits (76), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 17/36 (47%), Positives = 21/36 (58%), Gaps = 0/36 (0%)
Query 66 LPQIFYQIGPKFRDEARPRAGCLRAREFVMLEVYSF 101
+P Q+G FR+E PR G LR REF M E+ F
Sbjct 357 MPFAAAQLGLGFRNEIAPRNGLLRVREFQMGEIEHF 392
> pfa:PFL0670c bifunctional aminoacyl-tRNA synthetase, putative
(EC:6.1.1.15); K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=746
Score = 32.3 bits (72), Expect = 0.35, Method: Composition-based stats.
Identities = 20/57 (35%), Positives = 27/57 (47%), Gaps = 0/57 (0%)
Query 46 PTSEEAAAWLVASNIRSHRDLPQIFYQIGPKFRDEARPRAGCLRAREFVMLEVYSFH 102
PTSE + IRS+RDLP Q R E + +R REF+ E ++ H
Sbjct 358 PTSETIMYSVFPKWIRSYRDLPLKLNQWNTVVRWEFKQPTPFIRTREFLWQEGHTAH 414
> sce:YHR020W Protein of unknown function that may interact with
ribosomes, based on co-purification experiments; has similarity
to proline-tRNA ligase; YHR020W is an essential gene
(EC:6.1.1.15); K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=688
Score = 31.2 bits (69), Expect = 0.73, Method: Composition-based stats.
Identities = 24/88 (27%), Positives = 36/88 (40%), Gaps = 4/88 (4%)
Query 19 QNSDRIRSFGPSLYRLQDRRGRQFFLP----PTSEEAAAWLVASNIRSHRDLPQIFYQIG 74
+ D + F P + + + P PTSE A ++S+RDLP Q
Sbjct 260 KEKDHVEGFAPEVAWVTRAGSSELEEPIAIRPTSETVMYPYYAKWVQSYRDLPLKLNQWN 319
Query 75 PKFRDEARPRAGCLRAREFVMLEVYSFH 102
R E + LR REF+ E ++ H
Sbjct 320 SVVRWEFKHPQPFLRTREFLWQEGHTAH 347
> tpv:TP01_0166 prolyl-tRNA synthetase; K01881 prolyl-tRNA synthetase
[EC:6.1.1.15]
Length=683
Score = 31.2 bits (69), Expect = 0.75, Method: Composition-based stats.
Identities = 20/57 (35%), Positives = 28/57 (49%), Gaps = 0/57 (0%)
Query 46 PTSEEAAAWLVASNIRSHRDLPQIFYQIGPKFRDEARPRAGCLRAREFVMLEVYSFH 102
PTSE + IRS+RDLP Q R E + +R+REF+ E ++ H
Sbjct 305 PTSETIMYPEFSKWIRSYRDLPLKLNQWNSVVRWEFKQPTPFIRSREFLWQEGHTAH 361
> ath:AT5G52520 OVA6; OVA6 (OVULE ABORTION 6); ATP binding / aminoacyl-tRNA
ligase/ nucleotide binding / proline-tRNA ligase;
K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=543
Score = 31.2 bits (69), Expect = 0.90, Method: Composition-based stats.
Identities = 26/103 (25%), Positives = 45/103 (43%), Gaps = 6/103 (5%)
Query 5 SPLSLPNIMPIHWLQN-SDRIRSFGPSLYRLQDRRGRQF----FLPPTSEEAAAWLVASN 59
S + P +P +++ + + F P L + G++ + PTSE +
Sbjct 123 SNMYFPQFIPYSFIEKEASHVEGFSPELALVTVGGGKELEEKLVVRPTSETIVNHMFTQW 182
Query 60 IRSHRDLPQIFYQIGPKFRDEARPRAGCLRAREFVMLEVYSFH 102
I S+RDLP + Q R E R + +R EF+ E ++ H
Sbjct 183 IHSYRDLPLMINQWANVTRWEMRTKP-FIRTLEFLWQEGHTAH 224
> dre:317731 ttna, fi20g08, si:busm1-258d18.1, si:dz258d18.1,
ttn, wu:fi04b11, wu:fi20g08; titin a; K12567 titin [EC:2.7.11.1]
Length=32757
Score = 30.4 bits (67), Expect = 1.3, Method: Composition-based stats.
Identities = 25/101 (24%), Positives = 47/101 (46%), Gaps = 10/101 (9%)
Query 9 LPNIMPIHWLQNSDRIRSFGPSLYRLQDRRGRQFF-LPPTSEEAAAWLVASNI-RSHRDL 66
LP+I+ WL+NSD I +++ +G+ F +P TS +AW A+ I ++ RD
Sbjct 1356 LPDIV---WLKNSDIISPHKYPHIKIEGAKGQAHFQIPQTSGSDSAWYTATAINKAGRDT 1412
Query 67 PQIFYQIGPKFRDEARPR-----AGCLRAREFVMLEVYSFH 102
+ + ++ + R G +A++ E+ H
Sbjct 1413 TRCRVNVEVEYTEPVPERRLIIPKGTYKAKDVAPPEIEPLH 1453
> mmu:107508 Eprs, 2410081F06Rik, 3010002K18Rik, C79379, Qprs;
glutamyl-prolyl-tRNA synthetase (EC:6.1.1.15 6.1.1.17); K14163
bifunctional glutamyl/prolyl-tRNA synthetase [EC:6.1.1.17
6.1.1.15]
Length=1512
Score = 30.4 bits (67), Expect = 1.3, Method: Composition-based stats.
Identities = 21/55 (38%), Positives = 26/55 (47%), Gaps = 0/55 (0%)
Query 46 PTSEEAAAWLVASNIRSHRDLPQIFYQIGPKFRDEARPRAGCLRAREFVMLEVYS 100
PTSE A ++SHRDLP Q R E + LR REF+ E +S
Sbjct 1120 PTSETVMYPAYAKWVQSHRDLPVRLNQWCNVVRWEFKHPQPFLRTREFLWQEGHS 1174
> hsa:2058 EPRS, DKFZp313B047, EARS, GLUPRORS, PARS, QARS, QPRS;
glutamyl-prolyl-tRNA synthetase (EC:6.1.1.15 6.1.1.17); K14163
bifunctional glutamyl/prolyl-tRNA synthetase [EC:6.1.1.17
6.1.1.15]
Length=1512
Score = 30.4 bits (67), Expect = 1.5, Method: Composition-based stats.
Identities = 21/55 (38%), Positives = 26/55 (47%), Gaps = 0/55 (0%)
Query 46 PTSEEAAAWLVASNIRSHRDLPQIFYQIGPKFRDEARPRAGCLRAREFVMLEVYS 100
PTSE A ++SHRDLP Q R E + LR REF+ E +S
Sbjct 1120 PTSETVMYPAYAKWVQSHRDLPIKLNQWCNVVRWEFKHPQPFLRTREFLWQEGHS 1174
> tpv:TP01_0161 seryl-tRNA synthetase; K01875 seryl-tRNA synthetase
[EC:6.1.1.11]
Length=585
Score = 29.3 bits (64), Expect = 2.9, Method: Composition-based stats.
Identities = 26/100 (26%), Positives = 41/100 (41%), Gaps = 7/100 (7%)
Query 9 LPNIMPIHWLQNSDRIRSFGPSLYRLQDRR---GRQFFLPPTSEEAAAWLVASNIRSHRD 65
+P I+ L+ + + F +LY+L D G +L PTSE + L
Sbjct 345 VPLIVTKSTLELTGHLPYFEDNLYKLDDSNVFNGETGYLIPTSEVSLLGLFKGGKFKANS 404
Query 66 LPQIFYQIGPKFRDEARPRA----GCLRAREFVMLEVYSF 101
LP+ F FR E + G +R +F +E+ F
Sbjct 405 LPKFFTAYSECFRKEIQDYGANARGLIRQHQFGKVELLCF 444
> xla:100158438 eprs; glutamyl-prolyl-tRNA synthetase; K14163
bifunctional glutamyl/prolyl-tRNA synthetase [EC:6.1.1.17 6.1.1.15]
Length=1499
Score = 29.3 bits (64), Expect = 3.2, Method: Composition-based stats.
Identities = 20/52 (38%), Positives = 24/52 (46%), Gaps = 0/52 (0%)
Query 46 PTSEEAAAWLVASNIRSHRDLPQIFYQIGPKFRDEARPRAGCLRAREFVMLE 97
PTSE A ++SHRDLP Q R E + LR REF+ E
Sbjct 1107 PTSETVMYPAYAKWVQSHRDLPIKLNQWCNVVRWEFKHPQPFLRTREFLWQE 1158
> xla:100158287 hypothetical protein LOC100158287; K01881 prolyl-tRNA
synthetase [EC:6.1.1.15]
Length=655
Score = 29.3 bits (64), Expect = 3.3, Method: Composition-based stats.
Identities = 20/52 (38%), Positives = 24/52 (46%), Gaps = 0/52 (0%)
Query 46 PTSEEAAAWLVASNIRSHRDLPQIFYQIGPKFRDEARPRAGCLRAREFVMLE 97
PTSE A ++SHRDLP Q R E + LR REF+ E
Sbjct 263 PTSETVMYPAYAKWVQSHRDLPIKLNQWCNVVRWEFKHPQPFLRTREFLWQE 314
> ath:AT5G26830 threonyl-tRNA synthetase / threonine--tRNA ligase
(THRRS) (EC:6.1.1.3); K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=709
Score = 29.3 bits (64), Expect = 3.3, Method: Composition-based stats.
Identities = 21/93 (22%), Positives = 40/93 (43%), Gaps = 2/93 (2%)
Query 10 PNIMPIHWLQNSDRIRSFGPSLYRLQDRRGRQFFLPPTSEEAAAWLVASNIRSHRDLPQI 69
PN+ + Q S ++ +++ + ++F L P + + +RS+R+LP
Sbjct 369 PNMYNMELWQTSGHADNYKDNMFTFNIEK-QEFGLKPMNCPGHCLIFQHRVRSYRELPMR 427
Query 70 FYQIGPKFRDEAR-PRAGCLRAREFVMLEVYSF 101
G R+EA +G R R F + + F
Sbjct 428 LADFGVLHRNEASGALSGLTRVRRFQQDDAHIF 460
> dre:562037 eprs, wu:fb38d08, wu:ft34d10; glutamyl-prolyl-tRNA
synthetase; K14163 bifunctional glutamyl/prolyl-tRNA synthetase
[EC:6.1.1.17 6.1.1.15]
Length=1511
Score = 29.3 bits (64), Expect = 3.5, Method: Composition-based stats.
Identities = 20/52 (38%), Positives = 24/52 (46%), Gaps = 0/52 (0%)
Query 46 PTSEEAAAWLVASNIRSHRDLPQIFYQIGPKFRDEARPRAGCLRAREFVMLE 97
PTSE A ++SHRDLP Q R E + LR REF+ E
Sbjct 1119 PTSETVMYPAYAKWVQSHRDLPIKLNQWCNVVRWEFKHPQPFLRTREFLWQE 1170
> tgo:TGME49_025270 hypothetical protein
Length=3219
Score = 27.7 bits (60), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 11/35 (31%), Positives = 20/35 (57%), Gaps = 0/35 (0%)
Query 38 RGRQFFLPPTSEEAAAWLVASNIRSHRDLPQIFYQ 72
RG+ FF P + + AA+W +S + +H P ++
Sbjct 390 RGKAFFSPSSPDTAASWGASSELSAHGRPPDEAHE 424
Lambda K H
0.327 0.139 0.442
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2031832220
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40