bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2170_orf1
Length=92
Score E
Sequences producing significant alignments: (Bits) Value
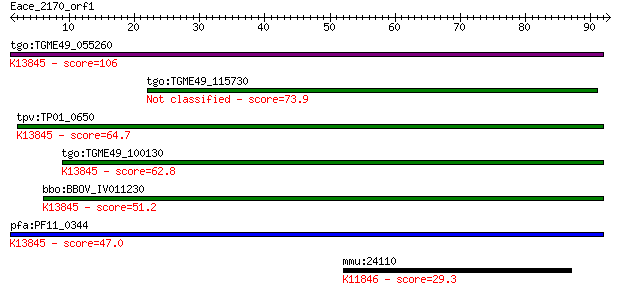
tgo:TGME49_055260 apical membrane antigen 1, putative ; K13845... 106 2e-23
tgo:TGME49_115730 apical membrane antigen, putative 73.9 1e-13
tpv:TP01_0650 apical membrane antigen 1; K13845 apical merozoi... 64.7 7e-11
tgo:TGME49_100130 apical membrane antigen, putative ; K13845 a... 62.8 3e-10
bbo:BBOV_IV011230 23.m06405; apical membrane antigen 1; K13845... 51.2 9e-07
pfa:PF11_0344 AMA1, AMA-1, Pf83, RMA-1, RMA1; apical membrane ... 47.0 1e-05
mmu:24110 Usp18, 1110058H21Rik, AW047653, UBP43, Ubp15; ubiqui... 29.3 3.0
> tgo:TGME49_055260 apical membrane antigen 1, putative ; K13845
apical merozoite antigen 1
Length=569
Score = 106 bits (264), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 48/95 (50%), Positives = 66/95 (69%), Gaps = 4/95 (4%)
Query 1 TSEFISPMTAAKLNSFSQLKAKTPLGRCAELSYKTTA----GRNSLYRYPFVYDTQKELC 56
+ + ISP L S +KA T LGRCAE ++KT A + + YRYPFVYD++K LC
Sbjct 167 SGQRISPFPMELLEKNSNIKASTDLGRCAEFAFKTVAMDKNNKATKYRYPFVYDSKKRLC 226
Query 57 YFLFIPMQRLMGERYCSYEGNPPDLTWYCFQPKKS 91
+ L++ MQ + G++YCS +G PPDLTWYCF+P+KS
Sbjct 227 HILYVSMQLMEGKKYCSVKGEPPDLTWYCFKPRKS 261
> tgo:TGME49_115730 apical membrane antigen, putative
Length=388
Score = 73.9 bits (180), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 36/74 (48%), Positives = 42/74 (56%), Gaps = 5/74 (6%)
Query 22 KTPLGRCAELSYKTTAGR-----NSLYRYPFVYDTQKELCYFLFIPMQRLMGERYCSYEG 76
KT +GRCA +Y T A S Y+YPFVYD CY L + Q L GE+YCS G
Sbjct 209 KTAIGRCALYAYSTIAVNPSTNYTSTYKYPFVYDAVSRKCYVLSVSAQLLKGEKYCSVNG 268
Query 77 NPPDLTWYCFQPKK 90
P LTW CF+P K
Sbjct 269 TPSGLTWACFEPVK 282
> tpv:TP01_0650 apical membrane antigen 1; K13845 apical merozoite
antigen 1
Length=785
Score = 64.7 bits (156), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 37/93 (39%), Positives = 53/93 (56%), Gaps = 4/93 (4%)
Query 2 SEFISPMTAAKLNSFSQLKAKTPLGRCAELSYKTTAG--RNSLYRYPFVYDTQKELCYFL 59
++ +SP++A L S++ K ++ L CAE S G RNS YRYPFVYD ++LCY L
Sbjct 336 NQLLSPISAQVLRSWN-YKHESDLSNCAEYSRNIVPGSNRNSKYRYPFVYDESEKLCYIL 394
Query 60 FIPMQRLMGERYCSYE-GNPPDLTWYCFQPKKS 91
+ PMQ G +YC + + + C P KS
Sbjct 395 YSPMQYNQGVKYCDKDSADEGTSSLACMYPDKS 427
> tgo:TGME49_100130 apical membrane antigen, putative ; K13845
apical merozoite antigen 1
Length=493
Score = 62.8 bits (151), Expect = 3e-10, Method: Composition-based stats.
Identities = 34/96 (35%), Positives = 51/96 (53%), Gaps = 13/96 (13%)
Query 9 TAAKLNSFSQLKAKT---PLGRCA---------ELSYKTTAGRNSLYRYPFVYDTQKELC 56
TA ++S++ AK LG C + S T + YRY FV+D +KE+C
Sbjct 163 TATGVSSYAVEHAKNIRDDLGHCIWWARMTSAHDTSSSATNSKEDYYRYAFVWDPKKEMC 222
Query 57 YFLFIPMQRLMGE-RYCSYEGNPPDLTWYCFQPKKS 91
+ +++ MQ + G YC + P+LTWYCF P+KS
Sbjct 223 HIMYLNMQEMTGAGTYCKRGDSGPNLTWYCFHPEKS 258
> bbo:BBOV_IV011230 23.m06405; apical membrane antigen 1; K13845
apical merozoite antigen 1
Length=605
Score = 51.2 bits (121), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 30/96 (31%), Positives = 49/96 (51%), Gaps = 10/96 (10%)
Query 6 SPMTAAKLNSFSQLKAK------TPLGRCAELSYKT--TAGRNSLYRYPFVYDTQKELCY 57
S +TAAKL+ S + + C+E + + + + YRYPFV+D+ ++CY
Sbjct 186 SSVTAAKLSPVSAKDLRRWGYEGNDVANCSEYASNLIPASDKTTKYRYPFVFDSDNQMCY 245
Query 58 FLFIPMQRLMGERYCSYEGNPPDLT--WYCFQPKKS 91
L+ +Q G RYC +G+ + T C +P KS
Sbjct 246 ILYSAIQYNQGNRYCDNDGSSEEGTSSLLCMKPYKS 281
> pfa:PF11_0344 AMA1, AMA-1, Pf83, RMA-1, RMA1; apical membrane
antigen 1, AMA1; K13845 apical merozoite antigen 1
Length=622
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 29/97 (29%), Positives = 49/97 (50%), Gaps = 7/97 (7%)
Query 1 TSEFISPMTAAKLNSFSQ----LKAKTPLGRCAELSYKT--TAGRNSLYRYPFVYDTQKE 54
T +SPMT ++ F + +K L C+ + +NS Y+YP VYD + +
Sbjct 186 TEPLMSPMTLDEMRHFYKDNKYVKNLDELTLCSRHAGNMIPDNDKNSNYKYPAVYDDKDK 245
Query 55 LCYFLFIPMQRLMGERYCSYEGNPPDLTWYCFQPKKS 91
C+ L+I Q G RYC+ + + + + +CF+P K
Sbjct 246 KCHILYIAAQENNGPRYCNKDESKRN-SMFCFRPAKD 281
> mmu:24110 Usp18, 1110058H21Rik, AW047653, UBP43, Ubp15; ubiquitin
specific peptidase 18 (EC:3.1.2.-); K11846 ubiquitin carboxyl-terminal
hydrolase 18/41 [EC:3.1.2.15]
Length=368
Score = 29.3 bits (64), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 15/38 (39%), Positives = 21/38 (55%), Gaps = 3/38 (7%)
Query 52 QKELCYFLFIPMQRL-MGE--RYCSYEGNPPDLTWYCF 86
Q E+ Y LF + + M + YC+Y NP D W+CF
Sbjct 293 QSEIHYELFAVIAHVGMADFGHYCAYIRNPVDGKWFCF 330
Lambda K H
0.321 0.136 0.434
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2003222032
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40