bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2151_orf2
Length=159
Score E
Sequences producing significant alignments: (Bits) Value
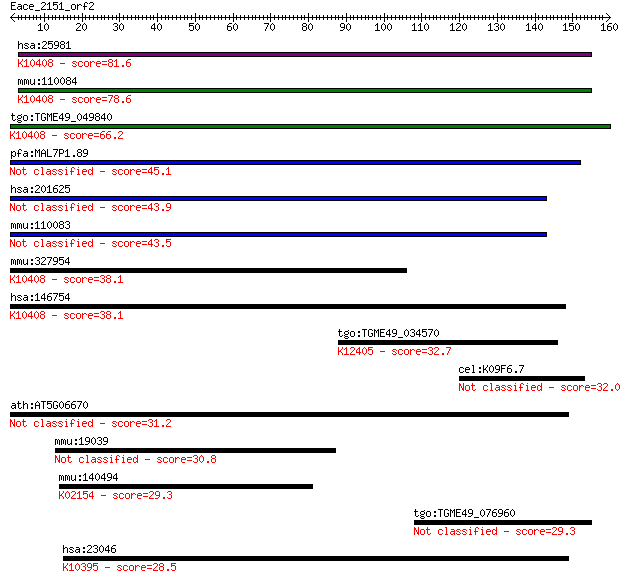
hsa:25981 DNAH1, DNAHC1, HDHC7, HL-11, HL11, XLHSRF-1; dynein,... 81.6 9e-16
mmu:110084 Dnahc1, B230373P09Rik, DKFZp434A236, Dnah1, E030034... 78.6 9e-15
tgo:TGME49_049840 dynein heavy chain domain containing protein... 66.2 5e-11
pfa:MAL7P1.89 dynein heavy chain, putative 45.1 9e-05
hsa:201625 DNAH12, DHC3, DLP12, DNAH12L, DNAH7L, DNAHC12, DNAH... 43.9 2e-04
mmu:110083 Dnahc12, 4921531P07Rik, DHC3, DLP12, Dnah12, Dnah12... 43.5 3e-04
mmu:327954 Dnahc2, 2900022L05Rik, 4930564A01, D130094J20, D330... 38.1 0.012
hsa:146754 DNAH2, DNAHC2, DNHD3, FLJ46675, KIAA1503; dynein, a... 38.1 0.014
tgo:TGME49_034570 peroxisomal multifunctional enzyme type 2, p... 32.7 0.53
cel:K09F6.7 hypothetical protein 32.0 0.92
ath:AT5G06670 ATP binding / microtubule motor 31.2 1.3
mmu:19039 Lgals3bp, 90K, CyCAP, MAC-2BP, Ppicap; lectin, galac... 30.8 1.9
mmu:140494 Atp6v0a4, Atp6n1b, a4; ATPase, H+ transporting, lys... 29.3 5.6
tgo:TGME49_076960 hypothetical protein 29.3 5.8
hsa:23046 KIF21B, FLJ16314; kinesin family member 21B; K10395 ... 28.5 9.4
> hsa:25981 DNAH1, DNAHC1, HDHC7, HL-11, HL11, XLHSRF-1; dynein,
axonemal, heavy chain 1; K10408 dynein heavy chain, axonemal
Length=4265
Score = 81.6 bits (200), Expect = 9e-16, Method: Compositional matrix adjust.
Identities = 47/154 (30%), Positives = 90/154 (58%), Gaps = 4/154 (2%)
Query 3 DLRRQLANKCTQITNSILEVLLQRVNLKVADVNDGFRAIRESLMKPTETPEEVVERNEFI 62
++++ L+ K + S+L++L + ++ +V + + FR+I + + + EE+ E E++
Sbjct 812 NVKQSLSKKRKALATSVLDILAKNLHKEVDSICEEFRSISRKIYEKPNSIEELAELREWM 871
Query 63 RMIPEKVFDLQTTIVEIQQINAILDTFQVPLSEDDFNRKWMAIGWPAQLDD--EIIRRQN 120
+ IPE++ L+ IV++ ++D F LS DDFN KW+A WP+++ E++++Q+
Sbjct 872 KGIPERLVGLEERIVKVMDDYQVMDEFLYNLSSDDFNDKWIASNWPSKILGQIELVQQQH 931
Query 121 ELEQTKRSLIVNMRKSQEAFQEKVSQMQLTVGGF 154
++ K I M Q FQEK+ +QL V GF
Sbjct 932 VEDEEKFRKIQIM--DQNNFQEKLEGLQLVVAGF 963
> mmu:110084 Dnahc1, B230373P09Rik, DKFZp434A236, Dnah1, E030034C22Rik,
MDHC7, MGC37121; dynein, axonemal, heavy chain 1;
K10408 dynein heavy chain, axonemal
Length=4250
Score = 78.6 bits (192), Expect = 9e-15, Method: Compositional matrix adjust.
Identities = 45/153 (29%), Positives = 88/153 (57%), Gaps = 2/153 (1%)
Query 3 DLRRQLANKCTQITNSILEVLLQRVNLKVADVNDGFRAIRESLMKPTETPEEVVERNEFI 62
++++ L+ K + S+L++L + ++ +V + + FR+I + + + EE+ E +++
Sbjct 797 NVKQSLSKKRKALATSMLDILAKNLHKEVDSICEEFRSISRKIYEKPNSIEELAELRDWM 856
Query 63 RMIPEKVFDLQTTIVEIQQINAILDTFQVPLSEDDFNRKWMAIGWPAQLDDEI-IRRQNE 121
+ IPEK+ L+ IV++ ++D F L+ DDFN KW A WP+++ +I + RQ
Sbjct 857 KGIPEKLVFLEERIVKVMSDYEVMDEFFYNLTTDDFNDKWAANNWPSKILGQIDMVRQQH 916
Query 122 LEQTKRSLIVNMRKSQEAFQEKVSQMQLTVGGF 154
+E ++ + + Q FQEK+ +QL V GF
Sbjct 917 VEDEEKFRKIQLM-DQNNFQEKLEGLQLVVAGF 948
> tgo:TGME49_049840 dynein heavy chain domain containing protein
; K10408 dynein heavy chain, axonemal
Length=4140
Score = 66.2 bits (160), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 44/159 (27%), Positives = 73/159 (45%), Gaps = 0/159 (0%)
Query 1 CTDLRRQLANKCTQITNSILEVLLQRVNLKVADVNDGFRAIRESLMKPTETPEEVVERNE 60
C D++R LA K + N IL+V+ QR + D FR +L K + EE+ E
Sbjct 707 CCDIKRFLAQKLHAVGNLILDVIAQRFRDQCTQTLDQFRGFYATLKKRPKNIEELTEMKT 766
Query 61 FIRMIPEKVFDLQTTIVEIQQINAILDTFQVPLSEDDFNRKWMAIGWPAQLDDEIIRRQN 120
FI IP K+ L I AIL+ F+ L +D N +W G P + + +
Sbjct 767 FIGDIPAKLERLAFDIKMNLHTFAILEEFKYKLYVEDHNLRWKMFGSPLETLTLMAETEK 826
Query 121 ELEQTKRSLIVNMRKSQEAFQEKVSQMQLTVGGFHRRTN 159
LE+ ++ + + Q F+E + ++ V F + ++
Sbjct 827 SLEKDRQVFLEELLTQQAEFEETIKDLEGIVSSFSQYSD 865
> pfa:MAL7P1.89 dynein heavy chain, putative
Length=5846
Score = 45.1 bits (105), Expect = 9e-05, Method: Composition-based stats.
Identities = 32/152 (21%), Positives = 71/152 (46%), Gaps = 2/152 (1%)
Query 1 CTDLRRQLANKCTQITNSILEVLLQRVNLKVADVNDGFRAIRESLMKPTETPEEVVERNE 60
C D++ + N+ ++ +++QR K + ++ L K T +E+ E+ E
Sbjct 673 CKDIKLFIINEINNYIKALTNIIVQRYKEKYTSNLMFYNSVIVRLKKKTTKIQEIYEKEE 732
Query 61 FIRMIPEKVFDLQTTIVEIQQINAILDTFQVPLSEDDFNRKWMAIGWPAQLDDEIIRRQN 120
+I+ + + + + I EI + L+ S DD+ W I P+++ ++I++ N
Sbjct 733 YIKDMKKSLDFVSQDIKEINILFNCLNKLNYKFSSDDYLSYWKIINRPSKI-EKIVKEVN 791
Query 121 E-LEQTKRSLIVNMRKSQEAFQEKVSQMQLTV 151
E +++ K L+ + + FQ + M+ V
Sbjct 792 ENIKKQKNVLLEELINDESKFQSSIVDMKENV 823
> hsa:201625 DNAH12, DHC3, DLP12, DNAH12L, DNAH7L, DNAHC12, DNAHC3,
DNHD2, FLJ40427, FLJ44290, HDHC3, HL-19, HL19; dynein,
axonemal, heavy chain 12
Length=3092
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 38/157 (24%), Positives = 71/157 (45%), Gaps = 19/157 (12%)
Query 1 CTDLRRQLANKCTQITNSILEVLLQRVNLKVADVNDGFRAIRESLMKPTETPEEVVERNE 60
C DL+ L NK N +L + + + + F AI+E +K ET EE+++
Sbjct 473 CEDLKTGLTNKAKAFANILLNDIASKYRKENECICSEFEAIKEHALKVPETTEEMMDLIS 532
Query 61 FIRMIPEKVFDLQTTIVEIQ----QINAILDTFQVPLSEDDFNRKWMAIGWPAQL----- 111
++ + ++ I+ IQ Q++ LD F P ++D + WP ++
Sbjct 533 YVEK--ARTVGIEELILRIQESKRQMSYFLDVFLFP--QEDLALNATVLMWPRKINPIFD 588
Query 112 -DDEII-----RRQNELEQTKRSLIVNMRKSQEAFQE 142
+DE+I +++NEL + LI+ + K +E
Sbjct 589 ENDELIENAKHKKENELMAKREKLILEIEKESRRMEE 625
> mmu:110083 Dnahc12, 4921531P07Rik, DHC3, DLP12, Dnah12, Dnah12l,
Dnahc3, Dnahc7c, Dnahc7l, Gm284, Gm74, Gm907, HL-19, HL19,
Hdhc3; dynein, axonemal, heavy chain 12
Length=985
Score = 43.5 bits (101), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 41/155 (26%), Positives = 71/155 (45%), Gaps = 15/155 (9%)
Query 1 CTDLRRQLANKCTQITNSILEVLLQRVNLKVADVNDGFRAIRESLMKPTETPEEVVERNE 60
C DL+ L NK N +L + + + + F AIR+ ++ ET EE++E
Sbjct 473 CEDLKIGLTNKARAFANILLNDIASKHRKENESICSEFEAIRDHALRVPETTEEMMELIA 532
Query 61 FI-RMIPEKVFDLQTTIVEIQ-QINAILDTFQVPLSEDDFNRKWMAIGWPAQL------D 112
F+ R + DL I E + Q++ LD +S++D N + WP ++ +
Sbjct 533 FVERARTVGILDLALRIQESKRQMSYFLDALL--MSQEDLNLNATVLLWPTKITPVFDEN 590
Query 113 DEIIR-----RQNELEQTKRSLIVNMRKSQEAFQE 142
DE+I ++NEL + LI+ + K +E
Sbjct 591 DELIENAKHAKENELIAKREKLILEIEKESRRMEE 625
> mmu:327954 Dnahc2, 2900022L05Rik, 4930564A01, D130094J20, D330014H01Rik,
Dnah2, Dnhd3; dynein, axonemal, heavy chain 2;
K10408 dynein heavy chain, axonemal
Length=4462
Score = 38.1 bits (87), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 23/111 (20%), Positives = 51/111 (45%), Gaps = 6/111 (5%)
Query 1 CTDLRRQLANKCTQITNSILEVLLQRVNLKVADVNDGFRAIRESLMKPTETPEEVVERNE 60
C+ L+ L C + N +L + ++AD++ + E + P +T EE+ +
Sbjct 1052 CSHLKFSLVQHCNEWQNKFTTLLKEMAAGRLADLHSYLKDNAEKISHPPQTLEELGVSLQ 1111
Query 61 FIRMIPEKVFDLQTTIVEIQQINAILDTFQVPLSE------DDFNRKWMAI 105
+ + + +L+T I I + IL+ ++VP+ + + N +W+
Sbjct 1112 LMDTLQHDLPNLETQIPPIHEQFTILEKYEVPVPDTVLEMLESLNGEWLTF 1162
> hsa:146754 DNAH2, DNAHC2, DNHD3, FLJ46675, KIAA1503; dynein,
axonemal, heavy chain 2; K10408 dynein heavy chain, axonemal
Length=4427
Score = 38.1 bits (87), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 27/153 (17%), Positives = 67/153 (43%), Gaps = 12/153 (7%)
Query 1 CTDLRRQLANKCTQITNSILEVLLQRVNLKVADVNDGFRAIRESLMKPTETPEEVVERNE 60
C+ L+ L C + N +L + ++ +++ + E + +P +T EE+ +
Sbjct 1017 CSHLKFSLVQHCNEWQNKFATLLREMAAGRLLELHTYLKENAEKISRPPQTLEELGVSLQ 1076
Query 61 FIRMIPEKVFDLQTTIVEIQQINAILDTFQVPLSE------DDFNRKWMAIGWPAQLDDE 114
+ + + +++T I I + AIL+ ++VP+ + D N +W+
Sbjct 1077 LVDALKHDLANVETQIPPIHEQFAILEKYEVPVEDSVLEMLDSLNGEWVV------FQQT 1130
Query 115 IIRRQNELEQTKRSLIVNMRKSQEAFQEKVSQM 147
++ + L++ K + S + F++K +
Sbjct 1131 LLDSKQMLKKHKEKFKTGLIHSADDFKKKAHTL 1163
> tgo:TGME49_034570 peroxisomal multifunctional enzyme type 2,
putative (EC:2.3.1.176 4.2.1.107); K12405 3-hydroxyacyl-CoA
dehydrogenase / 3a,7a,12a-trihydroxy-5b-cholest-24-enoyl-CoA
hydratase [EC:1.1.1.35 4.2.1.107]
Length=625
Score = 32.7 bits (73), Expect = 0.53, Method: Composition-based stats.
Identities = 20/67 (29%), Positives = 31/67 (46%), Gaps = 9/67 (13%)
Query 88 TFQVPLSEDDFNRKWMAI-------GWPAQLDDEIIRRQNELEQTKRSL--IVNMRKSQE 138
+F PLS DD R+W I +PA L D ++ +L+Q + R + +
Sbjct 254 SFATPLSPDDIAREWRHIRDFSGEVAYPASLQDSMLMVMQQLQQGTKPTDKRAEQRGASD 313
Query 139 AFQEKVS 145
A +EK S
Sbjct 314 AKEEKKS 320
> cel:K09F6.7 hypothetical protein
Length=299
Score = 32.0 bits (71), Expect = 0.92, Method: Compositional matrix adjust.
Identities = 13/33 (39%), Positives = 22/33 (66%), Gaps = 0/33 (0%)
Query 120 NELEQTKRSLIVNMRKSQEAFQEKVSQMQLTVG 152
+ L TK+SLI M KS+E Q +++++ T+G
Sbjct 127 SHLHHTKKSLIAEMEKSRELLQGRINRLNKTIG 159
> ath:AT5G06670 ATP binding / microtubule motor
Length=986
Score = 31.2 bits (69), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 33/151 (21%), Positives = 66/151 (43%), Gaps = 19/151 (12%)
Query 1 CTDLRRQLANKCTQITNSILEVLLQRVNLKVADVNDGFRAIRESLMKPTETPEEVVERNE 60
C DL+ ++AN Q++++ L++ D+N +++S P + E+V+E
Sbjct 752 CEDLQEEVANLKQQLSDA----------LELGDINSVTCHMQQSSQSPNKNEEKVIEAQA 801
Query 61 FIRMIPEKVFDLQTTIVEIQQINAILDTFQVPLSEDDFNRKWMAIGWPAQ---LDDEIIR 117
F ++ +L+ E+ ++N L+ L+E+ K +A + L +EI R
Sbjct 802 F------EIEELKLKAAELSELNEQLEIRNKKLAEESSYAKELASAAAIELKALSEEIAR 855
Query 118 RQNELEQTKRSLIVNMRKSQEAFQEKVSQMQ 148
N E+ L + S Q K ++
Sbjct 856 LMNHNERLAADLAAVQKSSVTTPQGKTGNLR 886
> mmu:19039 Lgals3bp, 90K, CyCAP, MAC-2BP, Ppicap; lectin, galactoside-binding,
soluble, 3 binding protein
Length=577
Score = 30.8 bits (68), Expect = 1.9, Method: Composition-based stats.
Identities = 18/77 (23%), Positives = 39/77 (50%), Gaps = 3/77 (3%)
Query 13 TQITNSILEVLLQRVNLKVADVNDGFRAIRE---SLMKPTETPEEVVERNEFIRMIPEKV 69
+ + ++++ LL + L V+ D +A+ + + E E +VE+ F M+P+++
Sbjct 297 SAVPTTLIQALLPKSELAVSSELDLLKAVDQWSTETIASHEDIERLVEQVRFPMMLPQEL 356
Query 70 FDLQTTIVEIQQINAIL 86
F+LQ + Q A+
Sbjct 357 FELQFNLSLYQDHQALF 373
> mmu:140494 Atp6v0a4, Atp6n1b, a4; ATPase, H+ transporting, lysosomal
V0 subunit A4 (EC:3.6.3.14); K02154 V-type H+-transporting
ATPase subunit I [EC:3.6.3.14]
Length=833
Score = 29.3 bits (64), Expect = 5.6, Method: Composition-based stats.
Identities = 20/69 (28%), Positives = 34/69 (49%), Gaps = 8/69 (11%)
Query 14 QITNSILEVLLQ--RVNLKVADVNDGFRAIRESLMKPTETPEEVVERNEFIRMIPEKVFD 71
+I +I + Q ++ LK+ + DGFRA + P PE ER E + + ++ D
Sbjct 216 EIKKNIFIIFYQGEQLRLKIKKICDGFRAT----IYP--CPEHAAERREMLTSVNVRLED 269
Query 72 LQTTIVEIQ 80
L T I + +
Sbjct 270 LITVITQTE 278
> tgo:TGME49_076960 hypothetical protein
Length=1779
Score = 29.3 bits (64), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 17/47 (36%), Positives = 25/47 (53%), Gaps = 6/47 (12%)
Query 108 PAQLDDEIIRRQNELEQTKRSLIVNMRKSQEAFQEKVSQMQLTVGGF 154
P + D+E IRR LE T ++ K EAF E+V ++ T+ G
Sbjct 17 PGKADEEAIRRLEALELTYKA------KEAEAFLERVEEVHETIQGL 57
> hsa:23046 KIF21B, FLJ16314; kinesin family member 21B; K10395
kinesin family member 4/7/21/27
Length=1624
Score = 28.5 bits (62), Expect = 9.4, Method: Compositional matrix adjust.
Identities = 39/146 (26%), Positives = 65/146 (44%), Gaps = 29/146 (19%)
Query 15 ITNSILEVLLQRVNLKVADVNDGFRAIRESLMKPTETPEEVVERNEFIRMIPEKVFDLQT 74
I N + +++ Q NL +A DG AI + +IR I E L+T
Sbjct 448 INNRVTQLMSQEANLLLAKAGDGNEAIGALI-------------QNYIREIEE----LRT 490
Query 75 TIVEIQQINAIL------DTFQVPLSEDDFNRKWMAIGWPA-QLDD--EIIRR-QNELEQ 124
++E + +N L + + P S G PA ++D E+IRR + +LE+
Sbjct 491 KLLESEAMNESLRRSLSRASARSPYSLGASPAAPAFGGSPASSMEDASEVIRRAKQDLER 550
Query 125 TKRSLIVNMRKS--QEAFQEKVSQMQ 148
K+ + RKS +EAF+++ Q
Sbjct 551 LKKKEVRQRRKSPEKEAFKKRAKLQQ 576
Lambda K H
0.320 0.134 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3647184800
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40